William Ogallo
Domain-agnostic and Multi-level Evaluation of Generative Models
Jan 20, 2023



Abstract:While the capabilities of generative models heavily improved in different domains (images, text, graphs, molecules, etc.), their evaluation metrics largely remain based on simplified quantities or manual inspection with limited practicality. To this end, we propose a framework for Multi-level Performance Evaluation of Generative mOdels (MPEGO), which could be employed across different domains. MPEGO aims to quantify generation performance hierarchically, starting from a sub-feature-based low-level evaluation to a global features-based high-level evaluation. MPEGO offers great customizability as the employed features are entirely user-driven and can thus be highly domain/problem-specific while being arbitrarily complex (e.g., outcomes of experimental procedures). We validate MPEGO using multiple generative models across several datasets from the material discovery domain. An ablation study is conducted to study the plausibility of intermediate steps in MPEGO. Results demonstrate that MPEGO provides a flexible, user-driven, and multi-level evaluation framework, with practical insights on the generation quality. The framework, source code, and experiments will be available at https://github.com/GT4SD/mpego.
A Collaborative Approach to the Analysis of the COVID-19 Response in Africa
Oct 04, 2022

Abstract:The COVID-19 crisis has emphasized the need for scientific methods such as machine learning to speed up the discovery of solutions to the pandemic. Harnessing machine learning techniques requires quality data, skilled personnel and advanced compute infrastructure. In Africa, however, machine learning competencies and compute infrastructures are limited. This paper demonstrates a cross-border collaborative capacity building approach to the application of machine learning techniques in discovering answers to COVID-19 questions.
Model-free feature selection to facilitate automatic discovery of divergent subgroups in tabular data
Mar 08, 2022



Abstract:Data-centric AI encourages the need of cleaning and understanding of data in order to achieve trustworthy AI. Existing technologies, such as AutoML, make it easier to design and train models automatically, but there is a lack of a similar level of capabilities to extract data-centric insights. Manual stratification of tabular data per a feature (e.g., gender) is limited to scale up for higher feature dimension, which could be addressed using automatic discovery of divergent subgroups. Nonetheless, these automatic discovery techniques often search across potentially exponential combinations of features that could be simplified using a preceding feature selection step. Existing feature selection techniques for tabular data often involve fitting a particular model in order to select important features. However, such model-based selection is prone to model-bias and spurious correlations in addition to requiring extra resource to design, fine-tune and train a model. In this paper, we propose a model-free and sparsity-based automatic feature selection (SAFS) framework to facilitate automatic discovery of divergent subgroups. Different from filter-based selection techniques, we exploit the sparsity of objective measures among feature values to rank and select features. We validated SAFS across two publicly available datasets (MIMIC-III and Allstate Claims) and compared it with six existing feature selection methods. SAFS achieves a reduction of feature selection time by a factor of 81x and 104x, averaged cross the existing methods in the MIMIC-III and Claims datasets respectively. SAFS-selected features are also shown to achieve competitive detection performance, e.g., 18.3% of features selected by SAFS in the Claims dataset detected divergent samples similar to those detected by using the whole features with a Jaccard similarity of 0.95 but with a 16x reduction in detection time.
Sparsity-based Feature Selection for Anomalous Subgroup Discovery
Jan 06, 2022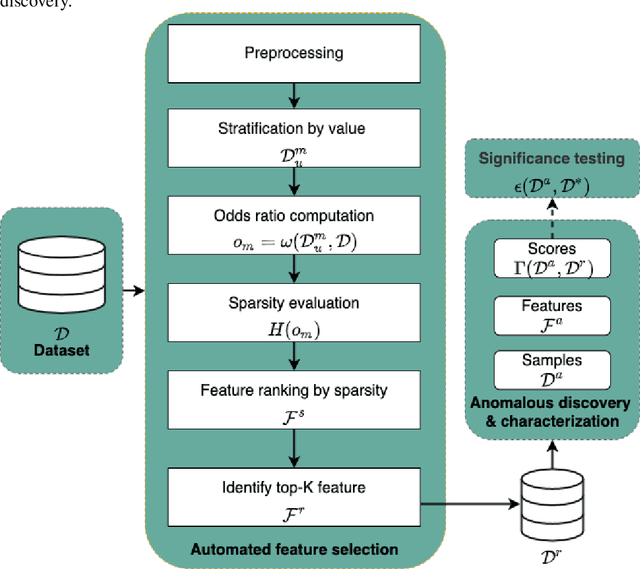
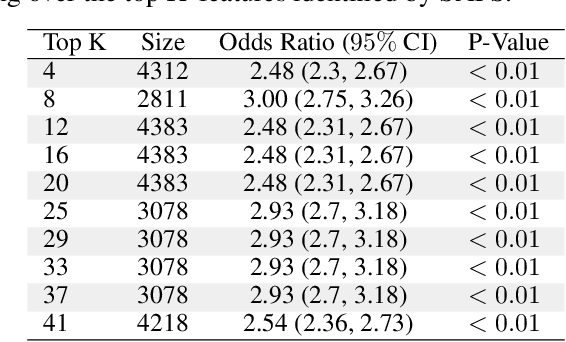
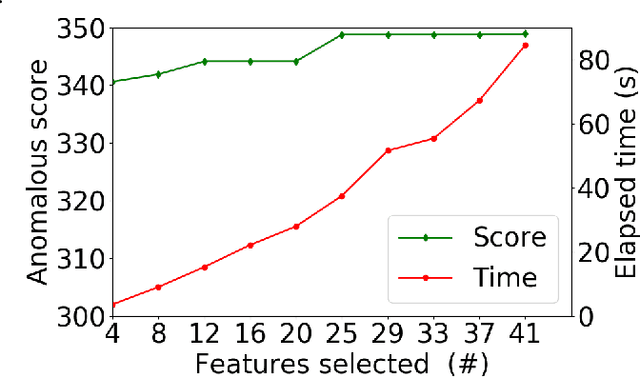
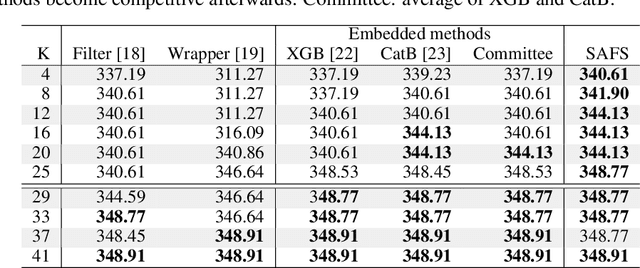
Abstract:Anomalous pattern detection aims to identify instances where deviation from normalcy is evident, and is widely applicable across domains. Multiple anomalous detection techniques have been proposed in the state of the art. However, there is a common lack of a principled and scalable feature selection method for efficient discovery. Existing feature selection techniques are often conducted by optimizing the performance of prediction outcomes rather than its systemic deviations from the expected. In this paper, we proposed a sparsity-based automated feature selection (SAFS) framework, which encodes systemic outcome deviations via the sparsity of feature-driven odds ratios. SAFS is a model-agnostic approach with usability across different discovery techniques. SAFS achieves more than $3\times$ reduction in computation time while maintaining detection performance when validated on publicly available critical care dataset. SAFS also results in a superior performance when compared against multiple baselines for feature selection.
Post-discovery Analysis of Anomalous Subsets
Nov 23, 2021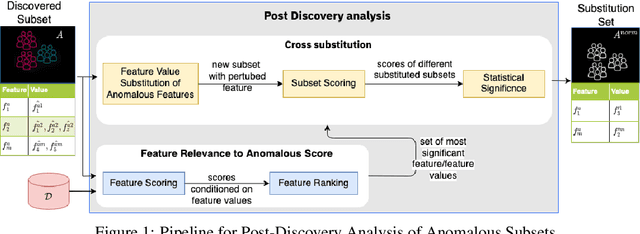
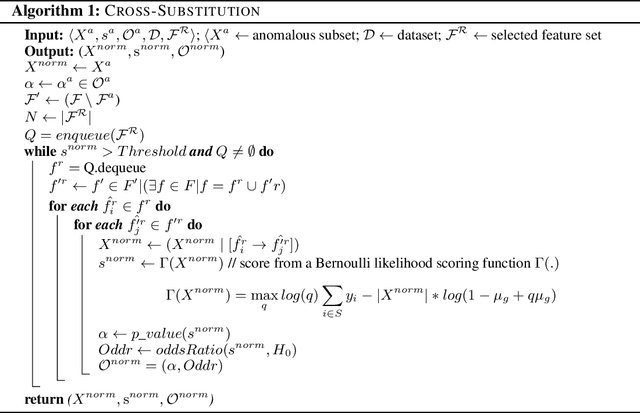
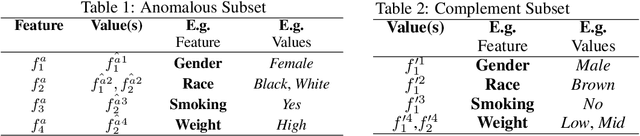
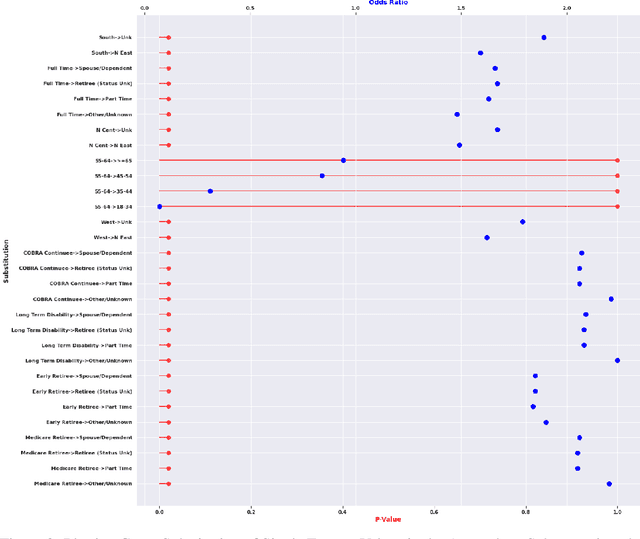
Abstract:Analyzing the behaviour of a population in response to disease and interventions is critical to unearth variability in healthcare as well as understand sub-populations that require specialized attention, but also to assist in designing future interventions. Two aspects become very essential in such analysis namely: i) Discovery of differentiating patterns exhibited by sub-populations, and ii) Characterization of the identified subpopulations. For the discovery phase, an array of approaches in the anomalous pattern detection literature have been employed to reveal differentiating patterns, especially to identify anomalous subgroups. However, these techniques are limited to describing the anomalous subgroups and offer little in form of insightful characterization, thereby limiting interpretability and understanding of these data-driven techniques in clinical practices. In this work, we propose an analysis of differentiated output (rather than discovery) and quantify anomalousness similarly to the counter-factual setting. To this end we design an approach to perform post-discovery analysis of anomalous subsets, in which we initially identify the most important features on the anomalousness of the subsets, then by perturbation, the approach seeks to identify the least number of changes necessary to lose anomalousness. Our approach is presented and the evaluation results on the 2019 MarketScan Commercial Claims and Medicare data, show that extra insights can be obtained by extrapolated examination of the identified subgroups.
Automated Supervised Feature Selection for Differentiated Patterns of Care
Nov 05, 2021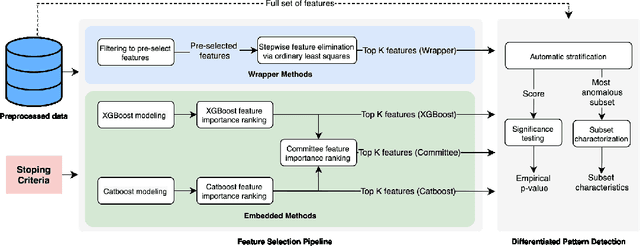
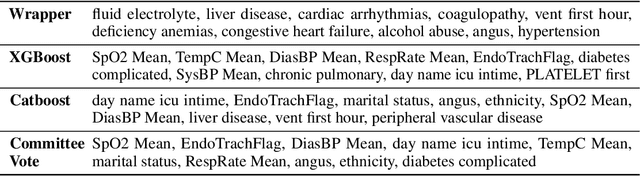
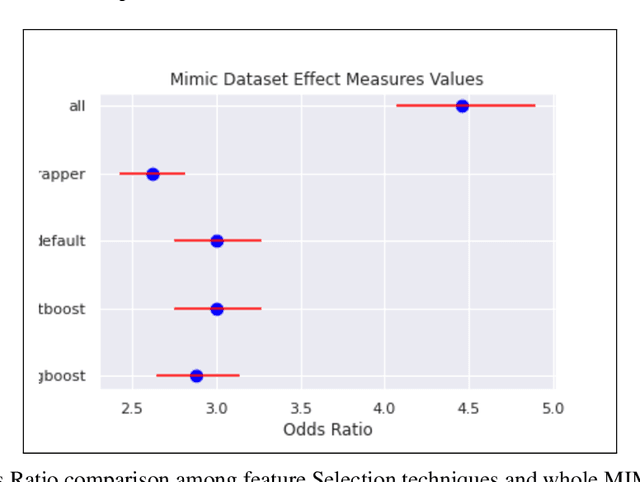
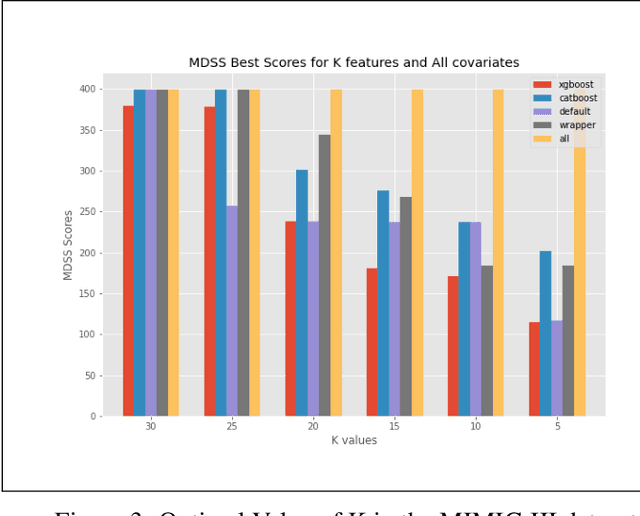
Abstract:An automated feature selection pipeline was developed using several state-of-the-art feature selection techniques to select optimal features for Differentiating Patterns of Care (DPOC). The pipeline included three types of feature selection techniques; Filters, Wrappers and Embedded methods to select the top K features. Five different datasets with binary dependent variables were used and their different top K optimal features selected. The selected features were tested in the existing multi-dimensional subset scanning (MDSS) where the most anomalous subpopulations, most anomalous subsets, propensity scores, and effect of measures were recorded to test their performance. This performance was compared with four similar metrics gained after using all covariates in the dataset in the MDSS pipeline. We found out that despite the different feature selection techniques used, the data distribution is key to note when determining the technique to use.
Disease Progression Modeling Workbench 360
Jun 24, 2021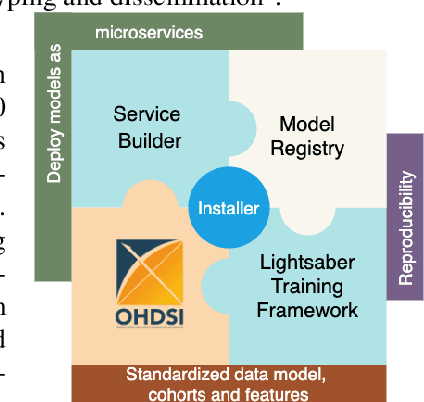
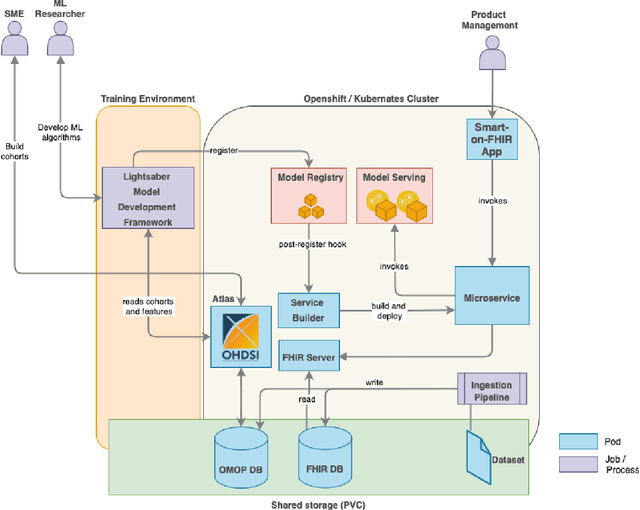
Abstract:In this work we introduce Disease Progression Modeling workbench 360 (DPM360) opensource clinical informatics framework for collaborative research and delivery of healthcare AI. DPM360, when fully developed, will manage the entire modeling life cycle, from data analysis (e.g., cohort identification) to machine learning algorithm development and prototyping. DPM360 augments the advantages of data model standardization and tooling (OMOP-CDM, Athena, ATLAS) provided by the widely-adopted OHDSI initiative with a powerful machine learning training framework, and a mechanism for rapid prototyping through automatic deployment of models as containerized services to a cloud environment.
WNTRAC: Artificial Intelligence Assisted Tracking of Non-pharmaceutical Interventions Implemented Worldwide for COVID-19
Sep 16, 2020



Abstract:The Coronavirus disease 2019 (COVID-19) global pandemic has transformed almost every facet of human society throughout the world. Against an emerging, highly transmissible disease with no definitive treatment or vaccine, governments worldwide have implemented non-pharmaceutical intervention (NPI) to slow the spread of the virus. Examples of such interventions include community actions (e.g. school closures, restrictions on mass gatherings), individual actions (e.g. mask wearing, self-quarantine), and environmental actions (e.g. public facility cleaning). We present the Worldwide Non-pharmaceutical Interventions Tracker for COVID-19 (WNTRAC), a comprehensive dataset consisting of over 6,000 NPIs implemented worldwide since the start of the pandemic. WNTRAC covers NPIs implemented across 261 countries and territories, and classifies NPI measures into a taxonomy of sixteen NPI types. NPI measures are automatically extracted daily from Wikipedia articles using natural language processing techniques and manually validated to ensure accuracy and veracity. We hope that the dataset is valuable for policymakers, public health leaders, and researchers in modeling and analysis efforts for controlling the spread of COVID-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge