Matteo Manica
We Need Improved Data Curation and Attribution in AI for Scientific Discovery
Apr 03, 2025



Abstract:As the interplay between human-generated and synthetic data evolves, new challenges arise in scientific discovery concerning the integrity of the data and the stability of the models. In this work, we examine the role of synthetic data as opposed to that of real experimental data for scientific research. Our analyses indicate that nearly three-quarters of experimental datasets available on open-access platforms have relatively low adoption rates, opening new opportunities to enhance their discoverability and usability by automated methods. Additionally, we observe an increasing difficulty in distinguishing synthetic from real experimental data. We propose supplementing ongoing efforts in automating synthetic data detection by increasing the focus on watermarking real experimental data, thereby strengthening data traceability and integrity. Our estimates suggest that watermarking even less than half of the real world data generated annually could help sustain model robustness, while promoting a balanced integration of synthetic and human-generated content.
Multiscale Byte Language Models -- A Hierarchical Architecture for Causal Million-Length Sequence Modeling
Feb 20, 2025Abstract:Bytes form the basis of the digital world and thus are a promising building block for multimodal foundation models. Recently, Byte Language Models (BLMs) have emerged to overcome tokenization, yet the excessive length of bytestreams requires new architectural paradigms. Therefore, we present the Multiscale Byte Language Model (MBLM), a model-agnostic hierarchical decoder stack that allows training with context windows of $5$M bytes on single GPU in full model precision. We thoroughly examine MBLM's performance with Transformer and Mamba blocks on both unimodal and multimodal tasks. Our experiments demonstrate that hybrid architectures are efficient in handling extremely long byte sequences during training while achieving near-linear generational efficiency. To the best of our knowledge, we present the first evaluation of BLMs on visual Q\&A tasks and find that, despite serializing images and the absence of an encoder, a MBLM with pure next token prediction can match custom CNN-LSTM architectures with designated classification heads. We show that MBLMs exhibit strong adaptability in integrating diverse data representations, including pixel and image filestream bytes, underlining their potential toward omnimodal foundation models. Source code is publicly available at: https://github.com/ai4sd/multiscale-byte-lm
Transforming the Hybrid Cloud for Emerging AI Workloads
Nov 20, 2024



Abstract:This white paper, developed through close collaboration between IBM Research and UIUC researchers within the IIDAI Institute, envisions transforming hybrid cloud systems to meet the growing complexity of AI workloads through innovative, full-stack co-design approaches, emphasizing usability, manageability, affordability, adaptability, efficiency, and scalability. By integrating cutting-edge technologies such as generative and agentic AI, cross-layer automation and optimization, unified control plane, and composable and adaptive system architecture, the proposed framework addresses critical challenges in energy efficiency, performance, and cost-effectiveness. Incorporating quantum computing as it matures will enable quantum-accelerated simulations for materials science, climate modeling, and other high-impact domains. Collaborative efforts between academia and industry are central to this vision, driving advancements in foundation models for material design and climate solutions, scalable multimodal data processing, and enhanced physics-based AI emulators for applications like weather forecasting and carbon sequestration. Research priorities include advancing AI agentic systems, LLM as an Abstraction (LLMaaA), AI model optimization and unified abstractions across heterogeneous infrastructure, end-to-end edge-cloud transformation, efficient programming model, middleware and platform, secure infrastructure, application-adaptive cloud systems, and new quantum-classical collaborative workflows. These ideas and solutions encompass both theoretical and practical research questions, requiring coordinated input and support from the research community. This joint initiative aims to establish hybrid clouds as secure, efficient, and sustainable platforms, fostering breakthroughs in AI-driven applications and scientific discovery across academia, industry, and society.
Unifying Molecular and Textual Representations via Multi-task Language Modelling
Jan 29, 2023Abstract:The recent advances in neural language models have also been successfully applied to the field of chemistry, offering generative solutions for classical problems in molecular design and synthesis planning. These new methods have the potential to optimize laboratory operations and fuel a new era of data-driven automation in scientific discovery. However, specialized models are still typically required for each task, leading to the need for problem-specific fine-tuning and neglecting task interrelations. The main obstacle in this field is the lack of a unified representation between natural language and chemical representations, complicating and limiting human-machine interaction. Here, we propose a multi-domain, multi-task language model to solve a wide range of tasks in both the chemical and natural language domains. By leveraging multi-task learning, our model can handle chemical and natural language concurrently, without requiring expensive pre-training on single domains or task-specific models. Interestingly, sharing weights across domains remarkably improves our model when benchmarked against state-of-the-art baselines on single-domain and cross-domain tasks. In particular, sharing information across domains and tasks gives rise to large improvements in cross-domain tasks, the magnitude of which increase with scale, as measured by more than a dozen of relevant metrics. Our work suggests that such models can robustly and efficiently accelerate discovery in physical sciences by superseding problem-specific fine-tuning and enhancing human-model interactions.
Domain-agnostic and Multi-level Evaluation of Generative Models
Jan 20, 2023



Abstract:While the capabilities of generative models heavily improved in different domains (images, text, graphs, molecules, etc.), their evaluation metrics largely remain based on simplified quantities or manual inspection with limited practicality. To this end, we propose a framework for Multi-level Performance Evaluation of Generative mOdels (MPEGO), which could be employed across different domains. MPEGO aims to quantify generation performance hierarchically, starting from a sub-feature-based low-level evaluation to a global features-based high-level evaluation. MPEGO offers great customizability as the employed features are entirely user-driven and can thus be highly domain/problem-specific while being arbitrarily complex (e.g., outcomes of experimental procedures). We validate MPEGO using multiple generative models across several datasets from the material discovery domain. An ablation study is conducted to study the plausibility of intermediate steps in MPEGO. Results demonstrate that MPEGO provides a flexible, user-driven, and multi-level evaluation framework, with practical insights on the generation quality. The framework, source code, and experiments will be available at https://github.com/GT4SD/mpego.
BLOOM: A 176B-Parameter Open-Access Multilingual Language Model
Nov 09, 2022Abstract:Large language models (LLMs) have been shown to be able to perform new tasks based on a few demonstrations or natural language instructions. While these capabilities have led to widespread adoption, most LLMs are developed by resource-rich organizations and are frequently kept from the public. As a step towards democratizing this powerful technology, we present BLOOM, a 176B-parameter open-access language model designed and built thanks to a collaboration of hundreds of researchers. BLOOM is a decoder-only Transformer language model that was trained on the ROOTS corpus, a dataset comprising hundreds of sources in 46 natural and 13 programming languages (59 in total). We find that BLOOM achieves competitive performance on a wide variety of benchmarks, with stronger results after undergoing multitask prompted finetuning. To facilitate future research and applications using LLMs, we publicly release our models and code under the Responsible AI License.
Z-BERT-A: a zero-shot Pipeline for Unknown Intent detection
Aug 18, 2022



Abstract:Intent discovery is a fundamental task in NLP, and it is increasingly relevant for a variety of industrial applications (Quarteroni 2018). The main challenge resides in the need to identify from input utterances novel unseen in-tents. Herein, we propose Z-BERT-A, a two-stage method for intent discovery relying on a Transformer architecture (Vaswani et al. 2017; Devlin et al. 2018), fine-tuned with Adapters (Pfeiffer et al. 2020), initially trained for Natural Language Inference (NLI), and later applied for unknown in-tent classification in a zero-shot setting. In our evaluation, we firstly analyze the quality of the model after adaptive fine-tuning on known classes. Secondly, we evaluate its performance casting intent classification as an NLI task. Lastly, we test the zero-shot performance of the model on unseen classes, showing how Z-BERT-A can effectively perform in-tent discovery by generating intents that are semantically similar, if not equal, to the ground truth ones. Our experiments show how Z-BERT-A is outperforming a wide variety of baselines in two zero-shot settings: known intents classification and unseen intent discovery. The proposed pipeline holds the potential to be widely applied in a variety of application for customer care. It enables automated dynamic triage using a lightweight model that, unlike large language models, can be easily deployed and scaled in a wide variety of business scenarios. Especially when considering a setting with limited hardware availability and performance whereon-premise or low resource cloud deployments are imperative. Z-BERT-A, predicting novel intents from a single utterance, represents an innovative approach for intent discovery, enabling online generation of novel intents. The pipeline is available as an installable python package at the following link: https://github.com/GT4SD/zberta.
GT4SD: Generative Toolkit for Scientific Discovery
Jul 08, 2022
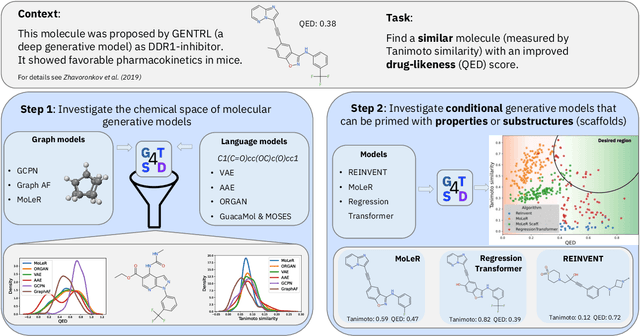
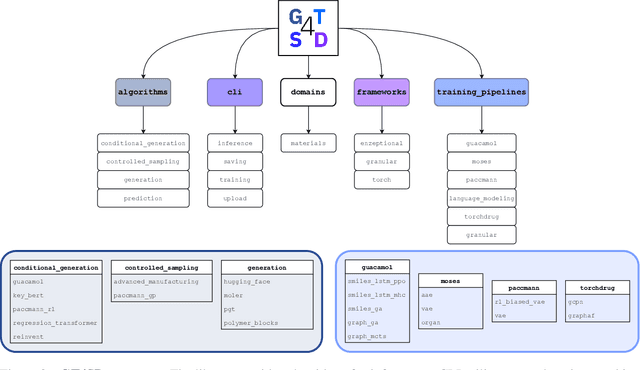
Abstract:With the growing availability of data within various scientific domains, generative models hold enormous potential to accelerate scientific discovery at every step of the scientific method. Perhaps their most valuable application lies in the speeding up of what has traditionally been the slowest and most challenging step of coming up with a hypothesis. Powerful representations are now being learned from large volumes of data to generate novel hypotheses, which is making a big impact on scientific discovery applications ranging from material design to drug discovery. The GT4SD (https://github.com/GT4SD/gt4sd-core) is an extensible open-source library that enables scientists, developers and researchers to train and use state-of-the-art generative models for hypothesis generation in scientific discovery. GT4SD supports a variety of uses of generative models across material science and drug discovery, including molecule discovery and design based on properties related to target proteins, omic profiles, scaffold distances, binding energies and more.
Regression Transformer: Concurrent Conditional Generation and Regression by Blending Numerical and Textual Tokens
Feb 01, 2022



Abstract:We report the Regression Transformer (RT), a method that abstracts regression as a conditional sequence modeling problem. The RT casts continuous properties as sequences of numerical tokens and encodes them jointly with conventional tokens. This yields a dichotomous model that can seamlessly transition between solving regression tasks and conditional generation tasks; solely governed by the mask location. We propose several extensions to the XLNet objective and adopt an alternating training scheme to concurrently optimize property prediction and conditional text generation based on a self-consistency loss. Our experiments on both chemical and protein languages demonstrate that the performance of traditional regression models can be surpassed despite training with cross entropy loss. Importantly, priming the same model with continuous properties yields a highly competitive conditional generative models that outperforms specialized approaches in a constrained property optimization benchmark. In sum, the Regression Transformer opens the door for "swiss army knife" models that excel at both regression and conditional generation. This finds application particularly in property-driven, local exploration of the chemical or protein space.
Multitask Prompted Training Enables Zero-Shot Task Generalization
Oct 15, 2021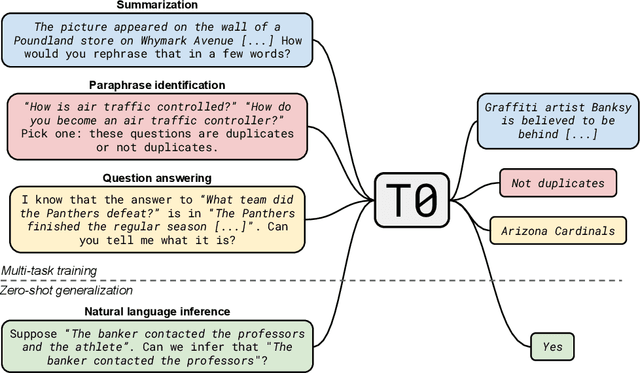

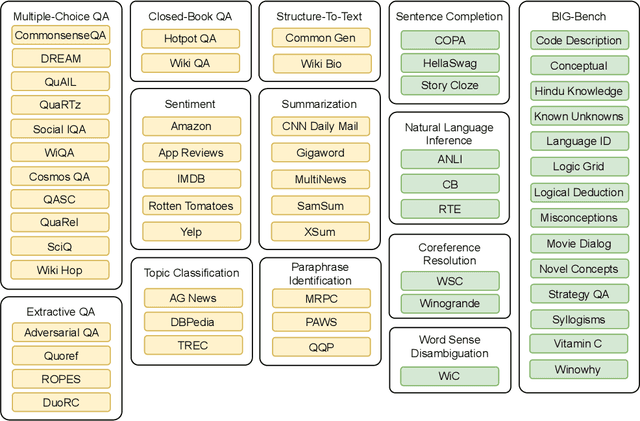

Abstract:Large language models have recently been shown to attain reasonable zero-shot generalization on a diverse set of tasks. It has been hypothesized that this is a consequence of implicit multitask learning in language model training. Can zero-shot generalization instead be directly induced by explicit multitask learning? To test this question at scale, we develop a system for easily mapping general natural language tasks into a human-readable prompted form. We convert a large set of supervised datasets, each with multiple prompts using varying natural language. These prompted datasets allow for benchmarking the ability of a model to perform completely unseen tasks specified in natural language. We fine-tune a pretrained encoder-decoder model on this multitask mixture covering a wide variety of tasks. The model attains strong zero-shot performance on several standard datasets, often outperforming models 16x its size. Further, our approach attains strong performance on a subset of tasks from the BIG-Bench benchmark, outperforming models 6x its size. All prompts and trained models are available at github.com/bigscience-workshop/promptsource/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge