Dimitrios Christofidellis
We Need Improved Data Curation and Attribution in AI for Scientific Discovery
Apr 03, 2025



Abstract:As the interplay between human-generated and synthetic data evolves, new challenges arise in scientific discovery concerning the integrity of the data and the stability of the models. In this work, we examine the role of synthetic data as opposed to that of real experimental data for scientific research. Our analyses indicate that nearly three-quarters of experimental datasets available on open-access platforms have relatively low adoption rates, opening new opportunities to enhance their discoverability and usability by automated methods. Additionally, we observe an increasing difficulty in distinguishing synthetic from real experimental data. We propose supplementing ongoing efforts in automating synthetic data detection by increasing the focus on watermarking real experimental data, thereby strengthening data traceability and integrity. Our estimates suggest that watermarking even less than half of the real world data generated annually could help sustain model robustness, while promoting a balanced integration of synthetic and human-generated content.
Unifying Molecular and Textual Representations via Multi-task Language Modelling
Jan 29, 2023Abstract:The recent advances in neural language models have also been successfully applied to the field of chemistry, offering generative solutions for classical problems in molecular design and synthesis planning. These new methods have the potential to optimize laboratory operations and fuel a new era of data-driven automation in scientific discovery. However, specialized models are still typically required for each task, leading to the need for problem-specific fine-tuning and neglecting task interrelations. The main obstacle in this field is the lack of a unified representation between natural language and chemical representations, complicating and limiting human-machine interaction. Here, we propose a multi-domain, multi-task language model to solve a wide range of tasks in both the chemical and natural language domains. By leveraging multi-task learning, our model can handle chemical and natural language concurrently, without requiring expensive pre-training on single domains or task-specific models. Interestingly, sharing weights across domains remarkably improves our model when benchmarked against state-of-the-art baselines on single-domain and cross-domain tasks. In particular, sharing information across domains and tasks gives rise to large improvements in cross-domain tasks, the magnitude of which increase with scale, as measured by more than a dozen of relevant metrics. Our work suggests that such models can robustly and efficiently accelerate discovery in physical sciences by superseding problem-specific fine-tuning and enhancing human-model interactions.
Z-BERT-A: a zero-shot Pipeline for Unknown Intent detection
Aug 18, 2022



Abstract:Intent discovery is a fundamental task in NLP, and it is increasingly relevant for a variety of industrial applications (Quarteroni 2018). The main challenge resides in the need to identify from input utterances novel unseen in-tents. Herein, we propose Z-BERT-A, a two-stage method for intent discovery relying on a Transformer architecture (Vaswani et al. 2017; Devlin et al. 2018), fine-tuned with Adapters (Pfeiffer et al. 2020), initially trained for Natural Language Inference (NLI), and later applied for unknown in-tent classification in a zero-shot setting. In our evaluation, we firstly analyze the quality of the model after adaptive fine-tuning on known classes. Secondly, we evaluate its performance casting intent classification as an NLI task. Lastly, we test the zero-shot performance of the model on unseen classes, showing how Z-BERT-A can effectively perform in-tent discovery by generating intents that are semantically similar, if not equal, to the ground truth ones. Our experiments show how Z-BERT-A is outperforming a wide variety of baselines in two zero-shot settings: known intents classification and unseen intent discovery. The proposed pipeline holds the potential to be widely applied in a variety of application for customer care. It enables automated dynamic triage using a lightweight model that, unlike large language models, can be easily deployed and scaled in a wide variety of business scenarios. Especially when considering a setting with limited hardware availability and performance whereon-premise or low resource cloud deployments are imperative. Z-BERT-A, predicting novel intents from a single utterance, represents an innovative approach for intent discovery, enabling online generation of novel intents. The pipeline is available as an installable python package at the following link: https://github.com/GT4SD/zberta.
GT4SD: Generative Toolkit for Scientific Discovery
Jul 08, 2022
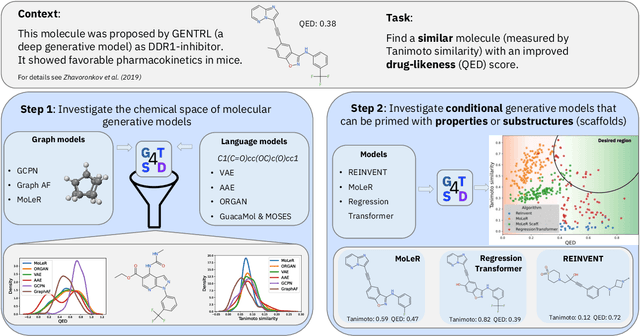
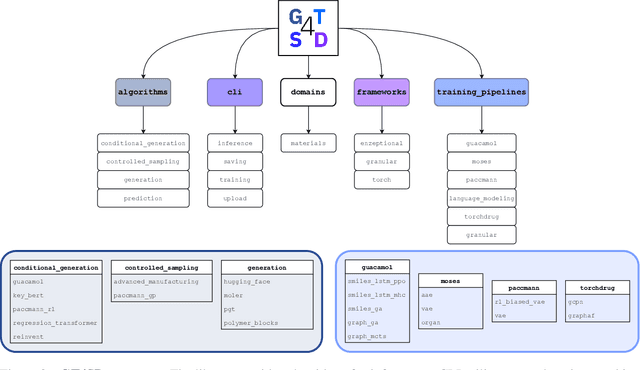
Abstract:With the growing availability of data within various scientific domains, generative models hold enormous potential to accelerate scientific discovery at every step of the scientific method. Perhaps their most valuable application lies in the speeding up of what has traditionally been the slowest and most challenging step of coming up with a hypothesis. Powerful representations are now being learned from large volumes of data to generate novel hypotheses, which is making a big impact on scientific discovery applications ranging from material design to drug discovery. The GT4SD (https://github.com/GT4SD/gt4sd-core) is an extensible open-source library that enables scientists, developers and researchers to train and use state-of-the-art generative models for hypothesis generation in scientific discovery. GT4SD supports a variety of uses of generative models across material science and drug discovery, including molecule discovery and design based on properties related to target proteins, omic profiles, scaffold distances, binding energies and more.
Understood in Translation, Transformers for Domain Understanding
Dec 18, 2020



Abstract:Knowledge acquisition is the essential first step of any Knowledge Graph (KG) application. This knowledge can be extracted from a given corpus (KG generation process) or specified from an existing KG (KG specification process). Focusing on domain specific solutions, knowledge acquisition is a labor intensive task usually orchestrated and supervised by subject matter experts. Specifically, the domain of interest is usually manually defined and then the needed generation or extraction tools are utilized to produce the KG. Herein, we propose a supervised machine learning method, based on Transformers, for domain definition of a corpus. We argue why such automated definition of the domain's structure is beneficial both in terms of construction time and quality of the generated graph. The proposed method is extensively validated on three public datasets (WebNLG, NYT and DocRED) by comparing it with two reference methods based on CNNs and RNNs models. The evaluation shows the efficiency of our model in this task. Focusing on scientific document understanding, we present a new health domain dataset based on publications extracted from PubMed and we successfully utilize our method on this. Lastly, we demonstrate how this work lays the foundation for fully automated and unsupervised KG generation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge