Ole Winther
Bi-Axial Transformers: Addressing the Increasing Complexity of EHR Classification
Aug 17, 2025Abstract:Electronic Health Records (EHRs), the digital representation of a patient's medical history, are a valuable resource for epidemiological and clinical research. They are also becoming increasingly complex, with recent trends indicating larger datasets, longer time series, and multi-modal integrations. Transformers, which have rapidly gained popularity due to their success in natural language processing and other domains, are well-suited to address these challenges due to their ability to model long-range dependencies and process data in parallel. But their application to EHR classification remains limited by data representations, which can reduce performance or fail to capture informative missingness. In this paper, we present the Bi-Axial Transformer (BAT), which attends to both the clinical variable and time point axes of EHR data to learn richer data relationships and address the difficulties of data sparsity. BAT achieves state-of-the-art performance on sepsis prediction and is competitive to top methods for mortality classification. In comparison to other transformers, BAT demonstrates increased robustness to data missingness, and learns unique sensor embeddings which can be used in transfer learning. Baseline models, which were previously located across multiple repositories or utilized deprecated libraries, were re-implemented with PyTorch and made available for reproduction and future benchmarking.
Generative Diffusion Models for Sequential Recommendations
Oct 25, 2024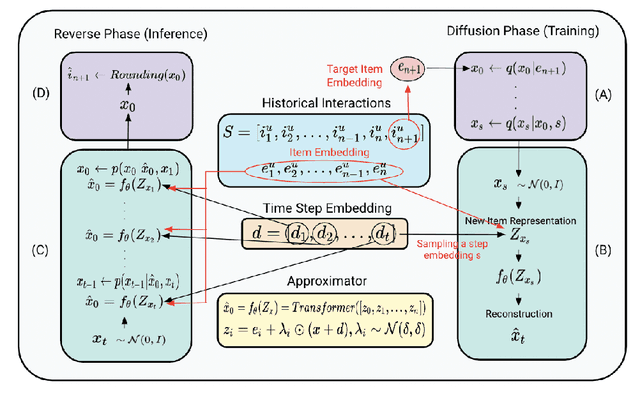
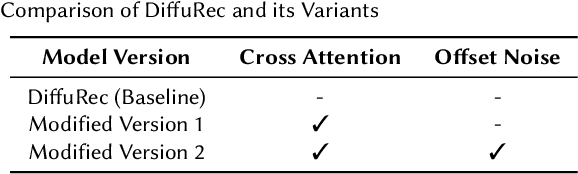
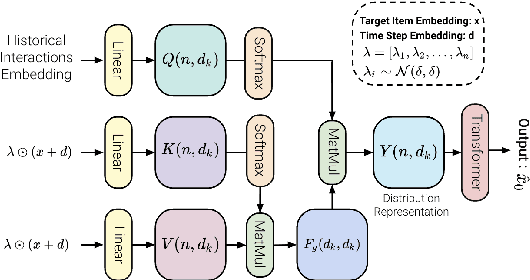
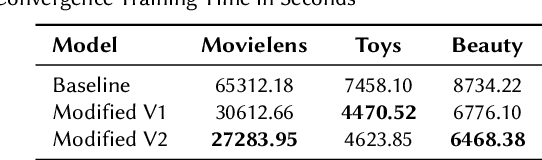
Abstract:Generative models such as Variational Autoencoders (VAEs) and Generative Adversarial Networks (GANs) have shown promise in sequential recommendation tasks. However, they face challenges, including posterior collapse and limited representation capacity. The work by Li et al. (2023) introduces a novel approach that leverages diffusion models to address these challenges by representing item embeddings as distributions rather than fixed vectors. This approach allows for a more adaptive reflection of users' diverse interests and various item aspects. During the diffusion phase, the model converts the target item embedding into a Gaussian distribution by adding noise, facilitating the representation of sequential item distributions and the injection of uncertainty. An Approximator then processes this noisy item representation to reconstruct the target item. In the reverse phase, the model utilizes users' past interactions to reverse the noise and finalize the item prediction through a rounding operation. This research introduces enhancements to the DiffuRec architecture, particularly by adding offset noise in the diffusion process to improve robustness and incorporating a cross-attention mechanism in the Approximator to better capture relevant user-item interactions. These contributions led to the development of a new model, DiffuRecSys, which improves performance. Extensive experiments conducted on three public benchmark datasets demonstrate that these modifications enhance item representation, effectively capture diverse user preferences, and outperform existing baselines in sequential recommendation research.
Geometry Fidelity for Spherical Images
Jul 25, 2024



Abstract:Spherical or omni-directional images offer an immersive visual format appealing to a wide range of computer vision applications. However, geometric properties of spherical images pose a major challenge for models and metrics designed for ordinary 2D images. Here, we show that direct application of Fr\'echet Inception Distance (FID) is insufficient for quantifying geometric fidelity in spherical images. We introduce two quantitative metrics accounting for geometric constraints, namely Omnidirectional FID (OmniFID) and Discontinuity Score (DS). OmniFID is an extension of FID tailored to additionally capture field-of-view requirements of the spherical format by leveraging cubemap projections. DS is a kernel-based seam alignment score of continuity across borders of 2D representations of spherical images. In experiments, OmniFID and DS quantify geometry fidelity issues that are undetected by FID.
Coherent energy and force uncertainty in deep learning force fields
Dec 07, 2023
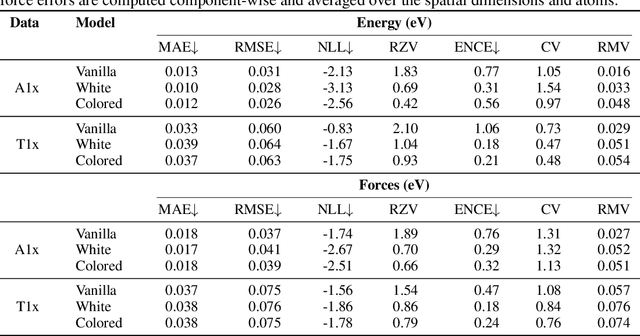


Abstract:In machine learning energy potentials for atomic systems, forces are commonly obtained as the negative derivative of the energy function with respect to atomic positions. To quantify aleatoric uncertainty in the predicted energies, a widely used modeling approach involves predicting both a mean and variance for each energy value. However, this model is not differentiable under the usual white noise assumption, so energy uncertainty does not naturally translate to force uncertainty. In this work we propose a machine learning potential energy model in which energy and force aleatoric uncertainty are linked through a spatially correlated noise process. We demonstrate our approach on an equivariant messages passing neural network potential trained on energies and forces on two out-of-equilibrium molecular datasets. Furthermore, we also show how to obtain epistemic uncertainties in this setting based on a Bayesian interpretation of deep ensemble models.
BEND: Benchmarking DNA Language Models on biologically meaningful tasks
Nov 25, 2023Abstract:The genome sequence contains the blueprint for governing cellular processes. While the availability of genomes has vastly increased over the last decades, experimental annotation of the various functional, non-coding and regulatory elements encoded in the DNA sequence remains both expensive and challenging. This has sparked interest in unsupervised language modeling of genomic DNA, a paradigm that has seen great success for protein sequence data. Although various DNA language models have been proposed, evaluation tasks often differ between individual works, and might not fully recapitulate the fundamental challenges of genome annotation, including the length, scale and sparsity of the data. In this study, we introduce BEND, a Benchmark for DNA language models, featuring a collection of realistic and biologically meaningful downstream tasks defined on the human genome. We find that embeddings from current DNA LMs can approach performance of expert methods on some tasks, but only capture limited information about long-range features. BEND is available at https://github.com/frederikkemarin/BEND.
DiffEnc: Variational Diffusion with a Learned Encoder
Oct 30, 2023Abstract:Diffusion models may be viewed as hierarchical variational autoencoders (VAEs) with two improvements: parameter sharing for the conditional distributions in the generative process and efficient computation of the loss as independent terms over the hierarchy. We consider two changes to the diffusion model that retain these advantages while adding flexibility to the model. Firstly, we introduce a data- and depth-dependent mean function in the diffusion process, which leads to a modified diffusion loss. Our proposed framework, DiffEnc, achieves state-of-the-art likelihood on CIFAR-10. Secondly, we let the ratio of the noise variance of the reverse encoder process and the generative process be a free weight parameter rather than being fixed to 1. This leads to theoretical insights: For a finite depth hierarchy, the evidence lower bound (ELBO) can be used as an objective for a weighted diffusion loss approach and for optimizing the noise schedule specifically for inference. For the infinite-depth hierarchy, on the other hand, the weight parameter has to be 1 to have a well-defined ELBO.
Image-free Classifier Injection for Zero-Shot Classification
Aug 21, 2023
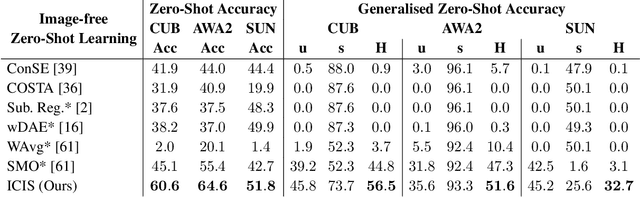

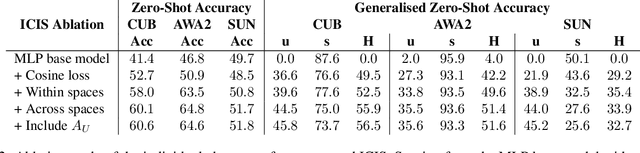
Abstract:Zero-shot learning models achieve remarkable results on image classification for samples from classes that were not seen during training. However, such models must be trained from scratch with specialised methods: therefore, access to a training dataset is required when the need for zero-shot classification arises. In this paper, we aim to equip pre-trained models with zero-shot classification capabilities without the use of image data. We achieve this with our proposed Image-free Classifier Injection with Semantics (ICIS) that injects classifiers for new, unseen classes into pre-trained classification models in a post-hoc fashion without relying on image data. Instead, the existing classifier weights and simple class-wise descriptors, such as class names or attributes, are used. ICIS has two encoder-decoder networks that learn to reconstruct classifier weights from descriptors (and vice versa), exploiting (cross-)reconstruction and cosine losses to regularise the decoding process. Notably, ICIS can be cheaply trained and applied directly on top of pre-trained classification models. Experiments on benchmark ZSL datasets show that ICIS produces unseen classifier weights that achieve strong (generalised) zero-shot classification performance. Code is available at https://github.com/ExplainableML/ImageFreeZSL .
Addressing caveats of neural persistence with deep graph persistence
Jul 20, 2023
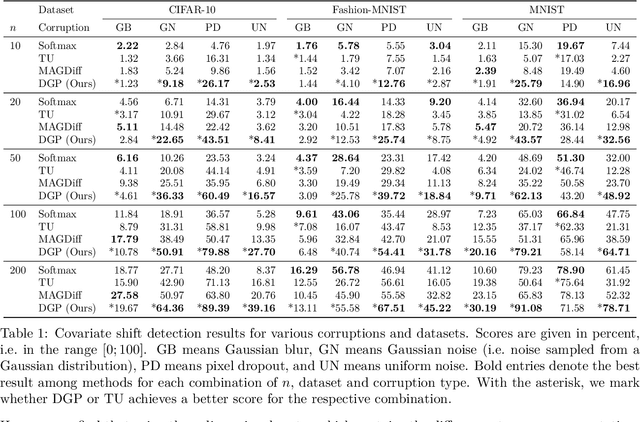
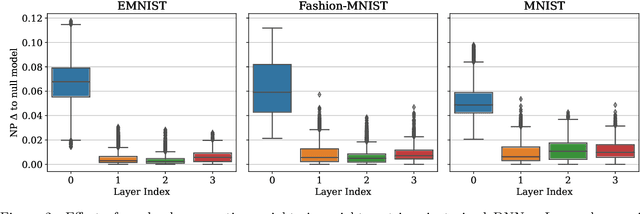
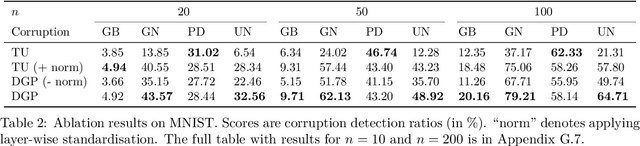
Abstract:Neural Persistence is a prominent measure for quantifying neural network complexity, proposed in the emerging field of topological data analysis in deep learning. In this work, however, we find both theoretically and empirically that the variance of network weights and spatial concentration of large weights are the main factors that impact neural persistence. Whilst this captures useful information for linear classifiers, we find that no relevant spatial structure is present in later layers of deep neural networks, making neural persistence roughly equivalent to the variance of weights. Additionally, the proposed averaging procedure across layers for deep neural networks does not consider interaction between layers. Based on our analysis, we propose an extension of the filtration underlying neural persistence to the whole neural network instead of single layers, which is equivalent to calculating neural persistence on one particular matrix. This yields our deep graph persistence measure, which implicitly incorporates persistent paths through the network and alleviates variance-related issues through standardisation. Code is available at https://github.com/ExplainableML/Deep-Graph-Persistence .
Aligning Optimization Trajectories with Diffusion Models for Constrained Design Generation
May 29, 2023



Abstract:Generative models have had a profound impact on vision and language, paving the way for a new era of multimodal generative applications. While these successes have inspired researchers to explore using generative models in science and engineering to accelerate the design process and reduce the reliance on iterative optimization, challenges remain. Specifically, engineering optimization methods based on physics still outperform generative models when dealing with constrained environments where data is scarce and precision is paramount. To address these challenges, we introduce Diffusion Optimization Models (DOM) and Trajectory Alignment (TA), a learning framework that demonstrates the efficacy of aligning the sampling trajectory of diffusion models with the optimization trajectory derived from traditional physics-based methods. This alignment ensures that the sampling process remains grounded in the underlying physical principles. Our method allows for generating feasible and high-performance designs in as few as two steps without the need for expensive preprocessing, external surrogate models, or additional labeled data. We apply our framework to structural topology optimization, a fundamental problem in mechanical design, evaluating its performance on in- and out-of-distribution configurations. Our results demonstrate that TA outperforms state-of-the-art deep generative models on in-distribution configurations and halves the inference computational cost. When coupled with a few steps of optimization, it also improves manufacturability for out-of-distribution conditions. By significantly improving performance and inference efficiency, DOM enables us to generate high-quality designs in just a few steps and guide them toward regions of high performance and manufacturability, paving the way for the widespread application of generative models in large-scale data-driven design.
Implicit Transfer Operator Learning: Multiple Time-Resolution Surrogates for Molecular Dynamics
May 29, 2023Abstract:Computing properties of molecular systems rely on estimating expectations of the (unnormalized) Boltzmann distribution. Molecular dynamics (MD) is a broadly adopted technique to approximate such quantities. However, stable simulations rely on very small integration time-steps ($10^{-15}\,\mathrm{s}$), whereas convergence of some moments, e.g. binding free energy or rates, might rely on sampling processes on time-scales as long as $10^{-1}\, \mathrm{s}$, and these simulations must be repeated for every molecular system independently. Here, we present Implict Transfer Operator (ITO) Learning, a framework to learn surrogates of the simulation process with multiple time-resolutions. We implement ITO with denoising diffusion probabilistic models with a new SE(3) equivariant architecture and show the resulting models can generate self-consistent stochastic dynamics across multiple time-scales, even when the system is only partially observed. Finally, we present a coarse-grained CG-SE3-ITO model which can quantitatively model all-atom molecular dynamics using only coarse molecular representations. As such, ITO provides an important step towards multiple time- and space-resolution acceleration of MD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge