Dennis Madsen
BEND: Benchmarking DNA Language Models on biologically meaningful tasks
Nov 25, 2023Abstract:The genome sequence contains the blueprint for governing cellular processes. While the availability of genomes has vastly increased over the last decades, experimental annotation of the various functional, non-coding and regulatory elements encoded in the DNA sequence remains both expensive and challenging. This has sparked interest in unsupervised language modeling of genomic DNA, a paradigm that has seen great success for protein sequence data. Although various DNA language models have been proposed, evaluation tasks often differ between individual works, and might not fully recapitulate the fundamental challenges of genome annotation, including the length, scale and sparsity of the data. In this study, we introduce BEND, a Benchmark for DNA language models, featuring a collection of realistic and biologically meaningful downstream tasks defined on the human genome. We find that embeddings from current DNA LMs can approach performance of expert methods on some tasks, but only capture limited information about long-range features. BEND is available at https://github.com/frederikkemarin/BEND.
GiNGR: Generalized Iterative Non-Rigid Point Cloud and Surface Registration Using Gaussian Process Regression
Mar 18, 2022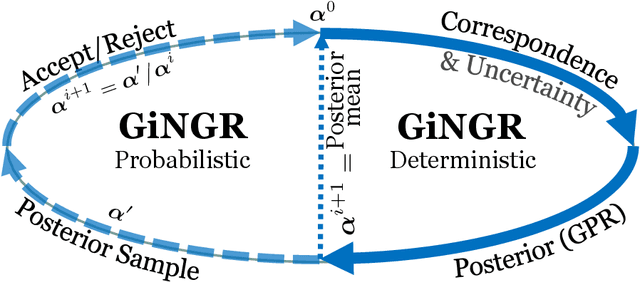



Abstract:In this paper, we unify popular non-rigid registration methods for point sets and surfaces under our general framework, GiNGR. GiNGR builds upon Gaussian Process Morphable Models (GPMM) and hence separates modeling the deformation prior from model adaptation for registration. In addition, it provides explainable hyperparameters, multi-resolution registration, trivial inclusion of expert annotation, and the ability to use and combine analytical and statistical deformation priors. But more importantly, the reformulation allows for a direct comparison of registration methods. Instead of using a general solver in the optimization step, we show how Gaussian process regression (GPR) iteratively can warp a reference onto a target, leading to smooth deformations following the prior for any dense, sparse, or partial estimated correspondences in a principled way. We show how the popular CPD and ICP algorithms can be directly explained with GiNGR. Furthermore, we show how existing algorithms in the GiNGR framework can perform probabilistic registration to obtain a distribution of different registrations instead of a single best registration. This can be used to analyze the uncertainty e.g. when registering partial observations. GiNGR is publicly available and fully modular to allow for domain-specific prior construction.
A Closest Point Proposal for MCMC-based Probabilistic Surface Registration
Jul 02, 2019



Abstract:In this paper, we propose a non-rigid surface registration algorithm that estimates the correspondence uncertainty using the Markov-chain Monte Carlo (MCMC) framework. The estimated uncertainty of the inferred registration is important for many applications, such as surgical planning or missing data reconstruction. The used Metropolis-Hastings (MH) algorithm decouples the inference from modelling the posterior using a propose-and-verify scheme. The widely used random sampling strategy leads to slow convergence rates in high dimensional space. In order to overcome this limitation, we introduce an informed probabilistic proposal based on ICP that can be used within the MH algorithm. While the ICP algorithm is used in the inference algorithm, the likelihood can be chosen independently. We showcase different surface distance measures, such as the traditional Euclidean norm and the Hausdorff distance. While quantifying the uncertainty of the correspondence, we also experimentally verify that our method is more robust than the non-rigid ICP algorithm and provides more accurate surface registrations. In a reconstruction task, we show how our probabilistic framework can be used to estimate the posterior distribution of missing data without assuming a fixed point-to-point correspondence. We have made our registration framework publicly available for the community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge