Antonio Foncubierta
We Need Improved Data Curation and Attribution in AI for Scientific Discovery
Apr 03, 2025



Abstract:As the interplay between human-generated and synthetic data evolves, new challenges arise in scientific discovery concerning the integrity of the data and the stability of the models. In this work, we examine the role of synthetic data as opposed to that of real experimental data for scientific research. Our analyses indicate that nearly three-quarters of experimental datasets available on open-access platforms have relatively low adoption rates, opening new opportunities to enhance their discoverability and usability by automated methods. Additionally, we observe an increasing difficulty in distinguishing synthetic from real experimental data. We propose supplementing ongoing efforts in automating synthetic data detection by increasing the focus on watermarking real experimental data, thereby strengthening data traceability and integrity. Our estimates suggest that watermarking even less than half of the real world data generated annually could help sustain model robustness, while promoting a balanced integration of synthetic and human-generated content.
BRACS: A Dataset for BReAst Carcinoma Subtyping in H&E Histology Images
Nov 08, 2021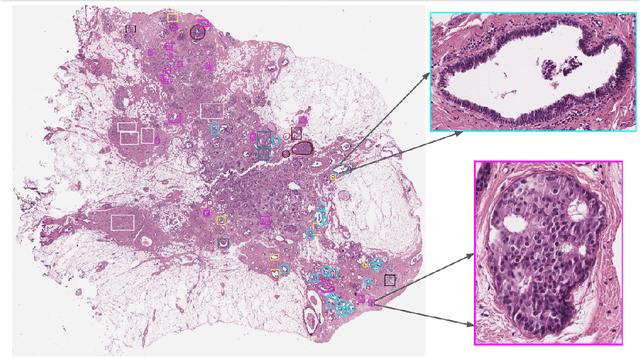

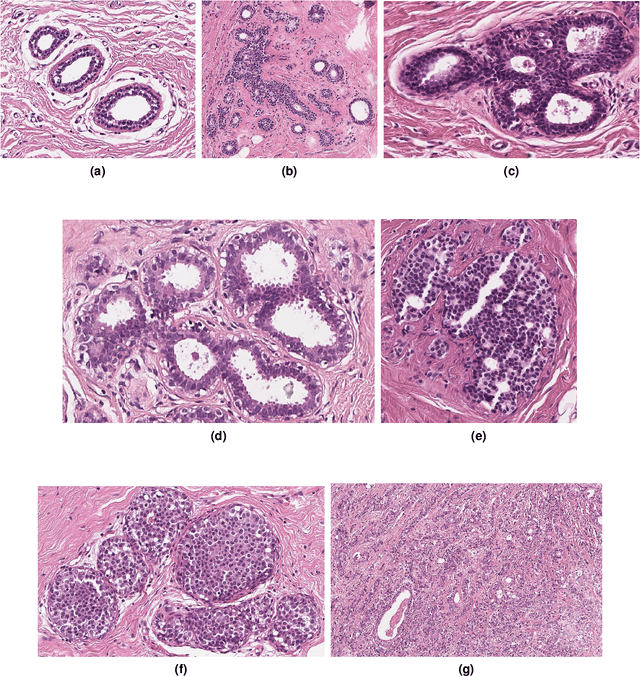
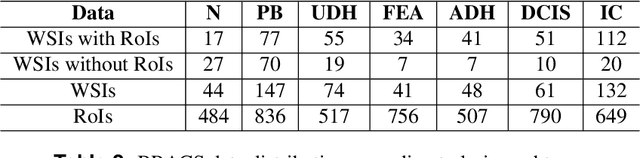
Abstract:Breast cancer is the most commonly diagnosed cancer and registers the highest number of deaths for women with cancer. Recent advancements in diagnostic activities combined with large-scale screening policies have significantly lowered the mortality rates for breast cancer patients. However, the manual inspection of tissue slides by the pathologists is cumbersome, time-consuming, and is subject to significant inter- and intra-observer variability. Recently, the advent of whole-slide scanning systems have empowered the rapid digitization of pathology slides, and enabled to develop digital workflows. These advances further enable to leverage Artificial Intelligence (AI) to assist, automate, and augment pathological diagnosis. But the AI techniques, especially Deep Learning (DL), require a large amount of high-quality annotated data to learn from. Constructing such task-specific datasets poses several challenges, such as, data-acquisition level constrains, time-consuming and expensive annotations, and anonymization of private information. In this paper, we introduce the BReAst Carcinoma Subtyping (BRACS) dataset, a large cohort of annotated Hematoxylin & Eosin (H&E)-stained images to facilitate the characterization of breast lesions. BRACS contains 547 Whole-Slide Images (WSIs), and 4539 Regions of Interest (ROIs) extracted from the WSIs. Each WSI, and respective ROIs, are annotated by the consensus of three board-certified pathologists into different lesion categories. Specifically, BRACS includes three lesion types, i.e., benign, malignant and atypical, which are further subtyped into seven categories. It is, to the best of our knowledge, the largest annotated dataset for breast cancer subtyping both at WSI- and ROI-level. Further, by including the understudied atypical lesions, BRACS offers an unique opportunity for leveraging AI to better understand their characteristics.
HistoCartography: A Toolkit for Graph Analytics in Digital Pathology
Jul 21, 2021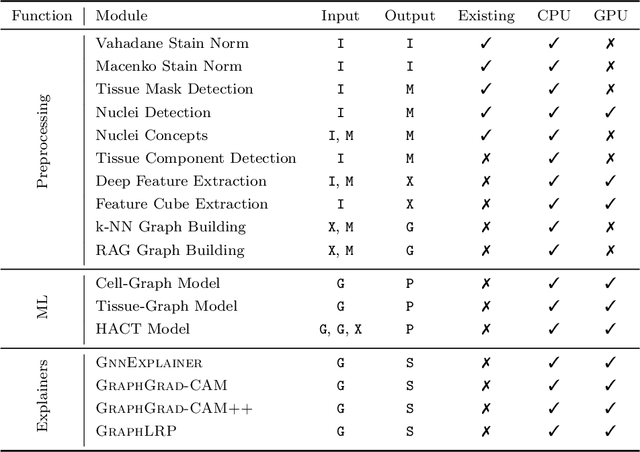
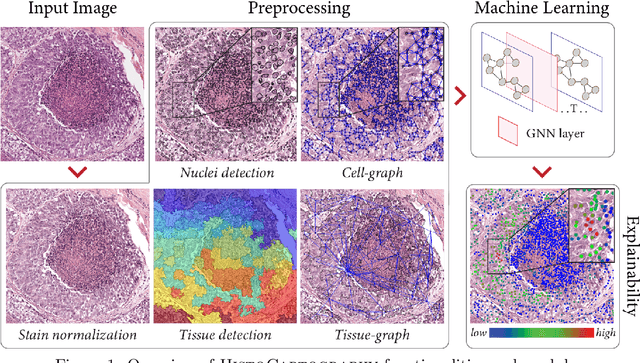
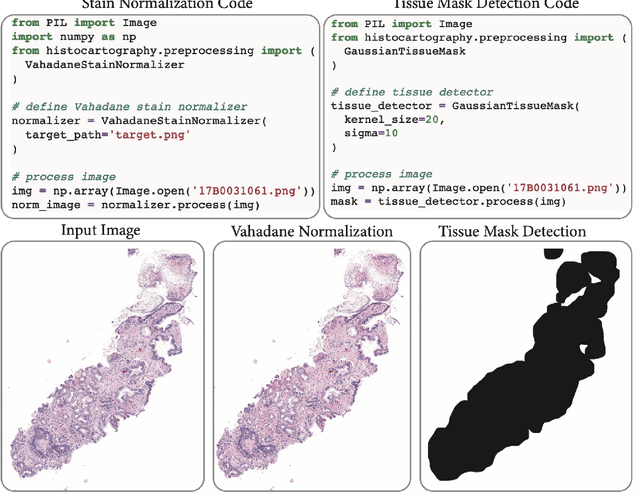
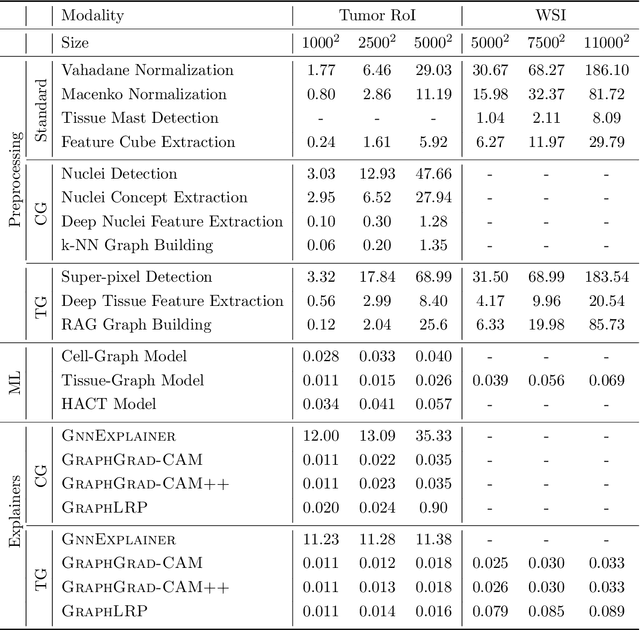
Abstract:Advances in entity-graph based analysis of histopathology images have brought in a new paradigm to describe tissue composition, and learn the tissue structure-to-function relationship. Entity-graphs offer flexible and scalable representations to characterize tissue organization, while allowing the incorporation of prior pathological knowledge to further support model interpretability and explainability. However, entity-graph analysis requires prerequisites for image-to-graph translation and knowledge of state-of-the-art machine learning algorithms applied to graph-structured data, which can potentially hinder their adoption. In this work, we aim to alleviate these issues by developing HistoCartography, a standardized python API with necessary preprocessing, machine learning and explainability tools to facilitate graph-analytics in computational pathology. Further, we have benchmarked the computational time and performance on multiple datasets across different imaging types and histopathology tasks to highlight the applicability of the API for building computational pathology workflows.
Hierarchical Graph Representations in Digital Pathology
Mar 17, 2021



Abstract:Cancer diagnosis, prognosis, and therapy response predictions from tissue specimens highly depend on the phenotype and topological distribution of constituting histological entities. Thus, adequate tissue representations for encoding histological entities is imperative for computer aided cancer patient care. To this end, several approaches have leveraged cell-graphs that encode cell morphology and organization to denote the tissue information. These allow for utilizing machine learning to map tissue representations to tissue functionality to help quantify their relationship. Though cellular information is crucial, it is incomplete alone to comprehensively characterize complex tissue structure. We herein treat the tissue as a hierarchical composition of multiple types of histological entities from fine to coarse level, capturing multivariate tissue information at multiple levels. We propose a novel multi-level hierarchical entity-graph representation of tissue specimens to model hierarchical compositions that encode histological entities as well as their intra- and inter-entity level interactions. Subsequently, a graph neural network is proposed to operate on the hierarchical entity-graph representation to map the tissue structure to tissue functionality. Specifically, for input histology images we utilize well-defined cells and tissue regions to build HierArchical Cell-to-Tissue (HACT) graph representations, and devise HACT-Net, a graph neural network, to classify such HACT representations. As part of this work, we introduce the BReAst Carcinoma Subtyping (BRACS) dataset, a large cohort of H&E stained breast tumor images, to evaluate our proposed methodology against pathologists and state-of-the-art approaches. Through comparative assessment and ablation studies, our method is demonstrated to yield superior classification results compared to alternative methods as well as pathologists.
A Canonical Architecture For Predictive Analytics on Longitudinal Patient Records
Jul 24, 2020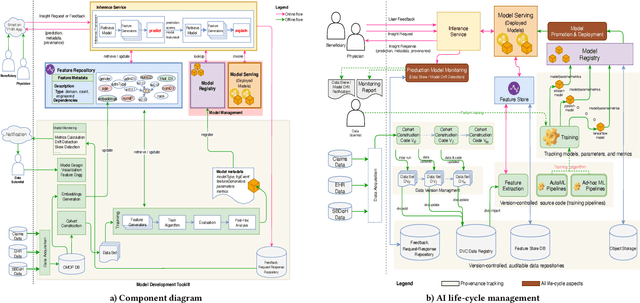
Abstract:Many institutions within the healthcare ecosystem are making significant investments in AI technologies to optimize their business operations at lower cost with improved patient outcomes. Despite the hype with AI, the full realization of this potential is seriously hindered by several systemic problems, including data privacy, security, bias, fairness, and explainability. In this paper, we propose a novel canonical architecture for the development of AI models in healthcare that addresses these challenges. This system enables the creation and management of AI predictive models throughout all the phases of their life cycle, including data ingestion, model building, and model promotion in production environments. This paper describes this architecture in detail, along with a qualitative evaluation of our experience of using it on real world problems.
HACT-Net: A Hierarchical Cell-to-Tissue Graph Neural Network for Histopathological Image Classification
Jul 01, 2020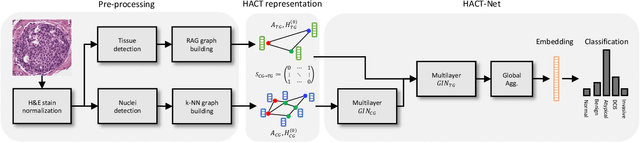
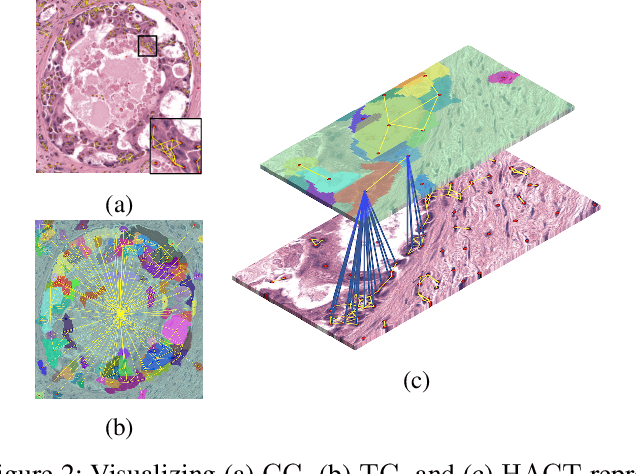
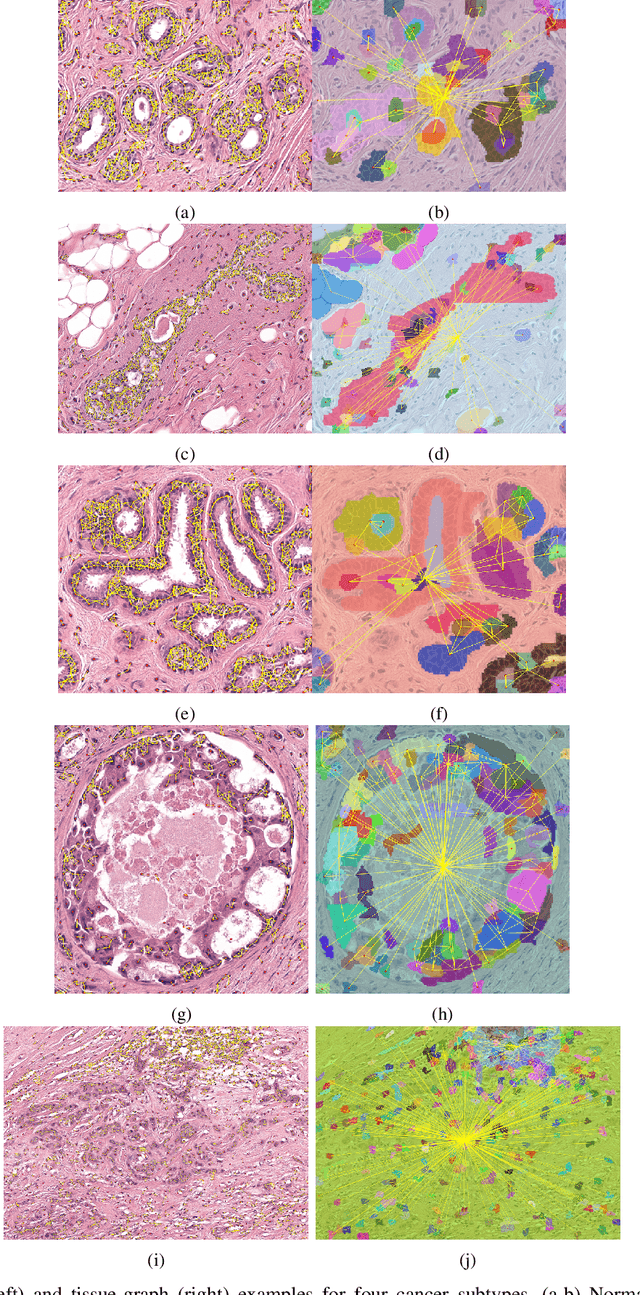

Abstract:Cancer diagnosis, prognosis, and therapeutic response prediction are heavily influenced by the relationship between the histopathological structures and the function of the tissue. Recent approaches acknowledging the structure-function relationship, have linked the structural and spatial patterns of cell organization in tissue via cell-graphs to tumor grades. Though cell organization is imperative, it is insufficient to entirely represent the histopathological structure. We propose a novel hierarchical cell-to-tissue-graph (HACT) representation to improve the structural depiction of the tissue. It consists of a low-level cell-graph, capturing cell morphology and interactions, a high-level tissue-graph, capturing morphology and spatial distribution of tissue parts, and cells-to-tissue hierarchies, encoding the relative spatial distribution of the cells with respect to the tissue distribution. Further, a hierarchical graph neural network (HACT-Net) is proposed to efficiently map the HACT representations to histopathological breast cancer subtypes. We assess the methodology on a large set of annotated tissue regions of interest from H\&E stained breast carcinoma whole-slides. Upon evaluation, the proposed method outperformed recent convolutional neural network and graph neural network approaches for breast cancer multi-class subtyping. The proposed entity-based topological analysis is more inline with the pathological diagnostic procedure of the tissue. It provides more command over the tissue modelling, therefore encourages the further inclusion of pathological priors into task-specific tissue representation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge