Pushpak Pati
TICON: A Slide-Level Tile Contextualizer for Histopathology Representation Learning
Dec 24, 2025Abstract:The interpretation of small tiles in large whole slide images (WSI) often needs a larger image context. We introduce TICON, a transformer-based tile representation contextualizer that produces rich, contextualized embeddings for ''any'' application in computational pathology. Standard tile encoder-based pipelines, which extract embeddings of tiles stripped from their context, fail to model the rich slide-level information essential for both local and global tasks. Furthermore, different tile-encoders excel at different downstream tasks. Therefore, a unified model is needed to contextualize embeddings derived from ''any'' tile-level foundation model. TICON addresses this need with a single, shared encoder, pretrained using a masked modeling objective to simultaneously unify and contextualize representations from diverse tile-level pathology foundation models. Our experiments demonstrate that TICON-contextualized embeddings significantly improve performance across many different tasks, establishing new state-of-the-art results on tile-level benchmarks (i.e., HEST-Bench, THUNDER, CATCH) and slide-level benchmarks (i.e., Patho-Bench). Finally, we pretrain an aggregator on TICON to form a slide-level foundation model, using only 11K WSIs, outperforming SoTA slide-level foundation models pretrained with up to 350K WSIs.
MEGAN: Mixture of Experts for Robust Uncertainty Estimation in Endoscopy Videos
Sep 16, 2025


Abstract:Reliable uncertainty quantification (UQ) is essential in medical AI. Evidential Deep Learning (EDL) offers a computationally efficient way to quantify model uncertainty alongside predictions, unlike traditional methods such as Monte Carlo (MC) Dropout and Deep Ensembles (DE). However, all these methods often rely on a single expert's annotations as ground truth for model training, overlooking the inter-rater variability in healthcare. To address this issue, we propose MEGAN, a Multi-Expert Gating Network that aggregates uncertainty estimates and predictions from multiple AI experts via EDL models trained with diverse ground truths and modeling strategies. MEGAN's gating network optimally combines predictions and uncertainties from each EDL model, enhancing overall prediction confidence and calibration. We extensively benchmark MEGAN on endoscopy videos for Ulcerative colitis (UC) disease severity estimation, assessed by visual labeling of Mayo Endoscopic Subscore (MES), where inter-rater variability is prevalent. In large-scale prospective UC clinical trial, MEGAN achieved a 3.5% improvement in F1-score and a 30.5% reduction in Expected Calibration Error (ECE) compared to existing methods. Furthermore, MEGAN facilitated uncertainty-guided sample stratification, reducing the annotation burden and potentially increasing efficiency and consistency in UC trials.
BioLangFusion: Multimodal Fusion of DNA, mRNA, and Protein Language Models
Jun 10, 2025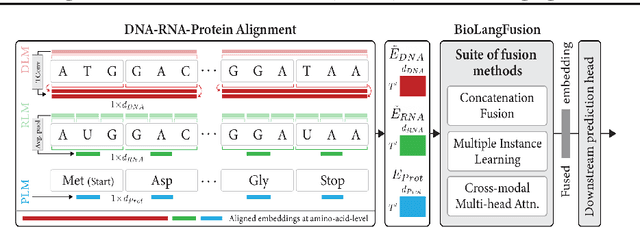
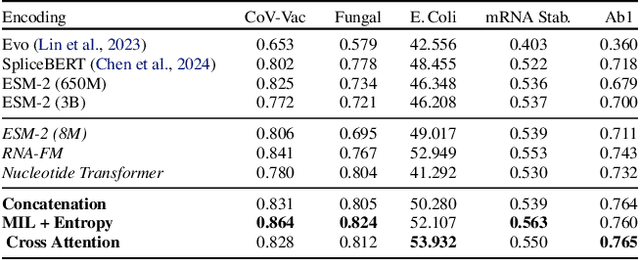
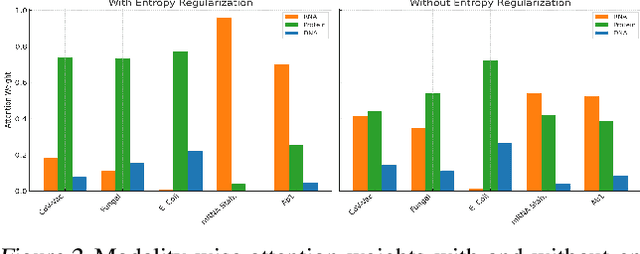
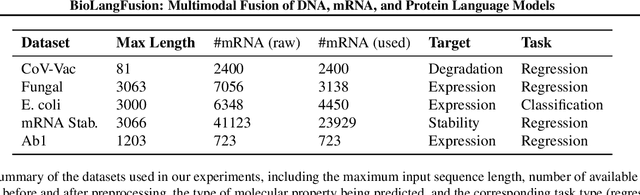
Abstract:We present BioLangFusion, a simple approach for integrating pre-trained DNA, mRNA, and protein language models into unified molecular representations. Motivated by the central dogma of molecular biology (information flow from gene to transcript to protein), we align per-modality embeddings at the biologically meaningful codon level (three nucleotides encoding one amino acid) to ensure direct cross-modal correspondence. BioLangFusion studies three standard fusion techniques: (i) codon-level embedding concatenation, (ii) entropy-regularized attention pooling inspired by multiple-instance learning, and (iii) cross-modal multi-head attention -- each technique providing a different inductive bias for combining modality-specific signals. These methods require no additional pre-training or modification of the base models, allowing straightforward integration with existing sequence-based foundation models. Across five molecular property prediction tasks, BioLangFusion outperforms strong unimodal baselines, showing that even simple fusion of pre-trained models can capture complementary multi-omic information with minimal overhead.
GECKO: Gigapixel Vision-Concept Contrastive Pretraining in Histopathology
Apr 01, 2025Abstract:Pretraining a Multiple Instance Learning (MIL) aggregator enables the derivation of Whole Slide Image (WSI)-level embeddings from patch-level representations without supervision. While recent multimodal MIL pretraining approaches leveraging auxiliary modalities have demonstrated performance gains over unimodal WSI pretraining, the acquisition of these additional modalities necessitates extensive clinical profiling. This requirement increases costs and limits scalability in existing WSI datasets lacking such paired modalities. To address this, we propose Gigapixel Vision-Concept Knowledge Contrastive pretraining (GECKO), which aligns WSIs with a Concept Prior derived from the available WSIs. First, we derive an inherently interpretable concept prior by computing the similarity between each WSI patch and textual descriptions of predefined pathology concepts. GECKO then employs a dual-branch MIL network: one branch aggregates patch embeddings into a WSI-level deep embedding, while the other aggregates the concept prior into a corresponding WSI-level concept embedding. Both aggregated embeddings are aligned using a contrastive objective, thereby pretraining the entire dual-branch MIL model. Moreover, when auxiliary modalities such as transcriptomics data are available, GECKO seamlessly integrates them. Across five diverse tasks, GECKO consistently outperforms prior unimodal and multimodal pretraining approaches while also delivering clinically meaningful interpretability that bridges the gap between computational models and pathology expertise. Code is made available at https://github.com/bmi-imaginelab/GECKO
Efficient Parameter Optimisation for Quantum Kernel Alignment: A Sub-sampling Approach in Variational Training
Jan 05, 2024



Abstract:Quantum machine learning with quantum kernels for classification problems is a growing area of research. Recently, quantum kernel alignment techniques that parameterise the kernel have been developed, allowing the kernel to be trained and therefore aligned with a specific dataset. While quantum kernel alignment is a promising technique, it has been hampered by considerable training costs because the full kernel matrix must be constructed at every training iteration. Addressing this challenge, we introduce a novel method that seeks to balance efficiency and performance. We present a sub-sampling training approach that uses a subset of the kernel matrix at each training step, thereby reducing the overall computational cost of the training. In this work, we apply the sub-sampling method to synthetic datasets and a real-world breast cancer dataset and demonstrate considerable reductions in the number of circuits required to train the quantum kernel while maintaining classification accuracy.
SI-MIL: Taming Deep MIL for Self-Interpretability in Gigapixel Histopathology
Dec 22, 2023



Abstract:Introducing interpretability and reasoning into Multiple Instance Learning (MIL) methods for Whole Slide Image (WSI) analysis is challenging, given the complexity of gigapixel slides. Traditionally, MIL interpretability is limited to identifying salient regions deemed pertinent for downstream tasks, offering little insight to the end-user (pathologist) regarding the rationale behind these selections. To address this, we propose Self-Interpretable MIL (SI-MIL), a method intrinsically designed for interpretability from the very outset. SI-MIL employs a deep MIL framework to guide an interpretable branch grounded on handcrafted pathological features, facilitating linear predictions. Beyond identifying salient regions, SI-MIL uniquely provides feature-level interpretations rooted in pathological insights for WSIs. Notably, SI-MIL, with its linear prediction constraints, challenges the prevalent myth of an inevitable trade-off between model interpretability and performance, demonstrating competitive results compared to state-of-the-art methods on WSI-level prediction tasks across three cancer types. In addition, we thoroughly benchmark the local- and global-interpretability of SI-MIL in terms of statistical analysis, a domain expert study, and desiderata of interpretability, namely, user-friendliness and faithfulness.
Hybrid quantum-classical graph neural networks for tumor classification in digital pathology
Oct 17, 2023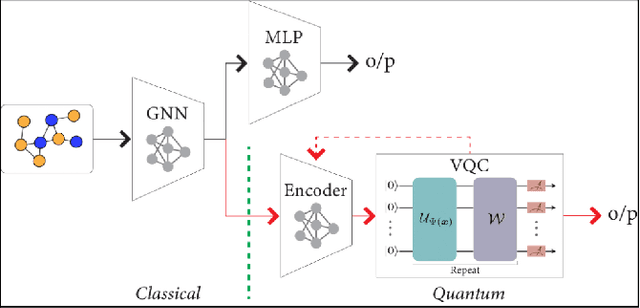
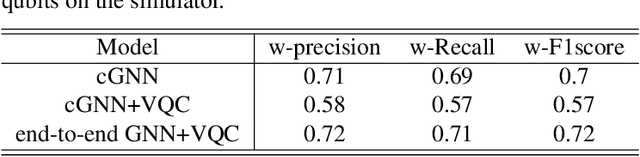
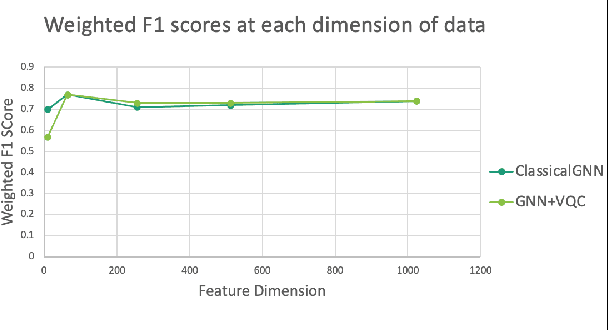
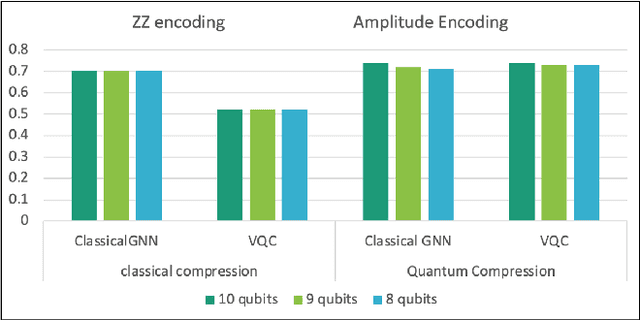
Abstract:Advances in classical machine learning and single-cell technologies have paved the way to understand interactions between disease cells and tumor microenvironments to accelerate therapeutic discovery. However, challenges in these machine learning methods and NP-hard problems in spatial Biology create an opportunity for quantum computing algorithms. We create a hybrid quantum-classical graph neural network (GNN) that combines GNN with a Variational Quantum Classifier (VQC) for classifying binary sub-tasks in breast cancer subtyping. We explore two variants of the same, the first with fixed pretrained GNN parameters and the second with end-to-end training of GNN+VQC. The results demonstrate that the hybrid quantum neural network (QNN) is at par with the state-of-the-art classical graph neural networks (GNN) in terms of weighted precision, recall and F1-score. We also show that by means of amplitude encoding, we can compress information in logarithmic number of qubits and attain better performance than using classical compression (which leads to information loss while keeping the number of qubits required constant in both regimes). Finally, we show that end-to-end training enables to improve over fixed GNN parameters and also slightly improves over vanilla GNN with same number of dimensions.
Multi-scale Feature Alignment for Continual Learning of Unlabeled Domains
Feb 02, 2023Abstract:Methods for unsupervised domain adaptation (UDA) help to improve the performance of deep neural networks on unseen domains without any labeled data. Especially in medical disciplines such as histopathology, this is crucial since large datasets with detailed annotations are scarce. While the majority of existing UDA methods focus on the adaptation from a labeled source to a single unlabeled target domain, many real-world applications with a long life cycle involve more than one target domain. Thus, the ability to sequentially adapt to multiple target domains becomes essential. In settings where the data from previously seen domains cannot be stored, e.g., due to data protection regulations, the above becomes a challenging continual learning problem. To this end, we propose to use generative feature-driven image replay in conjunction with a dual-purpose discriminator that not only enables the generation of images with realistic features for replay, but also promotes feature alignment during domain adaptation. We evaluate our approach extensively on a sequence of three histopathological datasets for tissue-type classification, achieving state-of-the-art results. We present detailed ablation experiments studying our proposed method components and demonstrate a possible use-case of our continual UDA method for an unsupervised patch-based segmentation task given high-resolution tissue images.
Weakly Supervised Joint Whole-Slide Segmentation and Classification in Prostate Cancer
Jan 07, 2023Abstract:The segmentation and automatic identification of histological regions of diagnostic interest offer a valuable aid to pathologists. However, segmentation methods are hampered by the difficulty of obtaining pixel-level annotations, which are tedious and expensive to obtain for Whole-Slide images (WSI). To remedy this, weakly supervised methods have been developed to exploit the annotations directly available at the image level. However, to our knowledge, none of these techniques is adapted to deal with WSIs. In this paper, we propose WholeSIGHT, a weakly-supervised method, to simultaneously segment and classify WSIs of arbitrary shapes and sizes. Formally, WholeSIGHT first constructs a tissue-graph representation of the WSI, where the nodes and edges depict tissue regions and their interactions, respectively. During training, a graph classification head classifies the WSI and produces node-level pseudo labels via post-hoc feature attribution. These pseudo labels are then used to train a node classification head for WSI segmentation. During testing, both heads simultaneously render class prediction and segmentation for an input WSI. We evaluated WholeSIGHT on three public prostate cancer WSI datasets. Our method achieved state-of-the-art weakly-supervised segmentation performance on all datasets while resulting in better or comparable classification with respect to state-of-the-art weakly-supervised WSI classification methods. Additionally, we quantify the generalization capability of our method in terms of segmentation and classification performance, uncertainty estimation, and model calibration.
Generative appearance replay for continual unsupervised domain adaptation
Jan 03, 2023



Abstract:Deep learning models can achieve high accuracy when trained on large amounts of labeled data. However, real-world scenarios often involve several challenges: Training data may become available in installments, may originate from multiple different domains, and may not contain labels for training. Certain settings, for instance medical applications, often involve further restrictions that prohibit retention of previously seen data due to privacy regulations. In this work, to address such challenges, we study unsupervised segmentation in continual learning scenarios that involve domain shift. To that end, we introduce GarDA (Generative Appearance Replay for continual Domain Adaptation), a generative-replay based approach that can adapt a segmentation model sequentially to new domains with unlabeled data. In contrast to single-step unsupervised domain adaptation (UDA), continual adaptation to a sequence of domains enables leveraging and consolidation of information from multiple domains. Unlike previous approaches in incremental UDA, our method does not require access to previously seen data, making it applicable in many practical scenarios. We evaluate GarDA on two datasets with different organs and modalities, where it substantially outperforms existing techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge