Pooya Mobadersany
Department of Pathology, Northwestern University, Chicago, IL, USA
MEGAN: Mixture of Experts for Robust Uncertainty Estimation in Endoscopy Videos
Sep 16, 2025


Abstract:Reliable uncertainty quantification (UQ) is essential in medical AI. Evidential Deep Learning (EDL) offers a computationally efficient way to quantify model uncertainty alongside predictions, unlike traditional methods such as Monte Carlo (MC) Dropout and Deep Ensembles (DE). However, all these methods often rely on a single expert's annotations as ground truth for model training, overlooking the inter-rater variability in healthcare. To address this issue, we propose MEGAN, a Multi-Expert Gating Network that aggregates uncertainty estimates and predictions from multiple AI experts via EDL models trained with diverse ground truths and modeling strategies. MEGAN's gating network optimally combines predictions and uncertainties from each EDL model, enhancing overall prediction confidence and calibration. We extensively benchmark MEGAN on endoscopy videos for Ulcerative colitis (UC) disease severity estimation, assessed by visual labeling of Mayo Endoscopic Subscore (MES), where inter-rater variability is prevalent. In large-scale prospective UC clinical trial, MEGAN achieved a 3.5% improvement in F1-score and a 30.5% reduction in Expected Calibration Error (ECE) compared to existing methods. Furthermore, MEGAN facilitated uncertainty-guided sample stratification, reducing the annotation burden and potentially increasing efficiency and consistency in UC trials.
Arges: Spatio-Temporal Transformer for Ulcerative Colitis Severity Assessment in Endoscopy Videos
Oct 01, 2024Abstract:Accurate assessment of disease severity from endoscopy videos in ulcerative colitis (UC) is crucial for evaluating drug efficacy in clinical trials. Severity is often measured by the Mayo Endoscopic Subscore (MES) and Ulcerative Colitis Endoscopic Index of Severity (UCEIS) score. However, expert MES/UCEIS annotation is time-consuming and susceptible to inter-rater variability, factors addressable by automation. Automation attempts with frame-level labels face challenges in fully-supervised solutions due to the prevalence of video-level labels in clinical trials. CNN-based weakly-supervised models (WSL) with end-to-end (e2e) training lack generalization to new disease scores and ignore spatio-temporal information crucial for accurate scoring. To address these limitations, we propose "Arges", a deep learning framework that utilizes a transformer with positional encoding to incorporate spatio-temporal information from frame features to estimate disease severity scores in endoscopy video. Extracted features are derived from a foundation model (ArgesFM), pre-trained on a large diverse dataset from multiple clinical trials (61M frames, 3927 videos). We evaluate four UC disease severity scores, including MES and three UCEIS component scores. Test set evaluation indicates significant improvements, with F1 scores increasing by 4.1% for MES and 18.8%, 6.6%, 3.8% for the three UCEIS component scores compared to state-of-the-art methods. Prospective validation on previously unseen clinical trial data further demonstrates the model's successful generalization.
NuCLS: A scalable crowdsourcing, deep learning approach and dataset for nucleus classification, localization and segmentation
Feb 18, 2021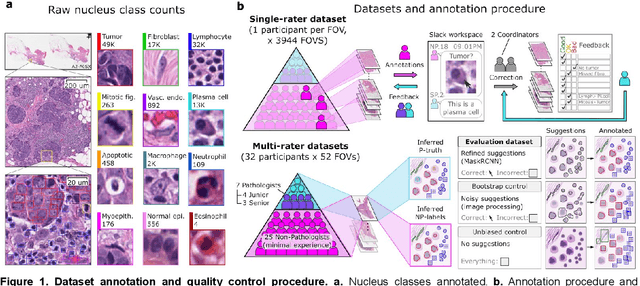
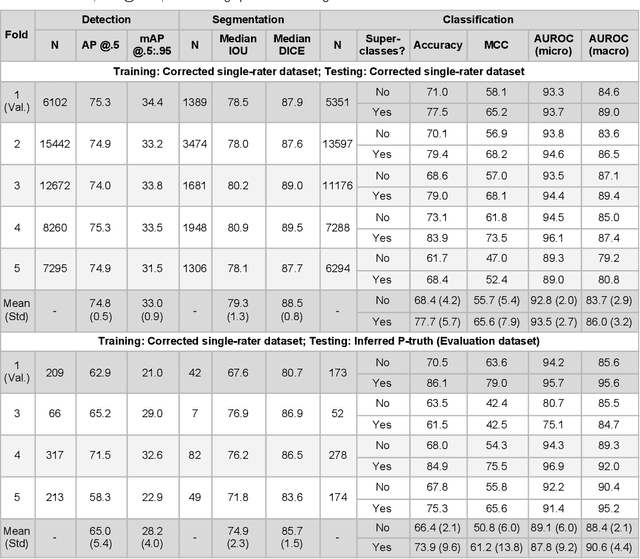
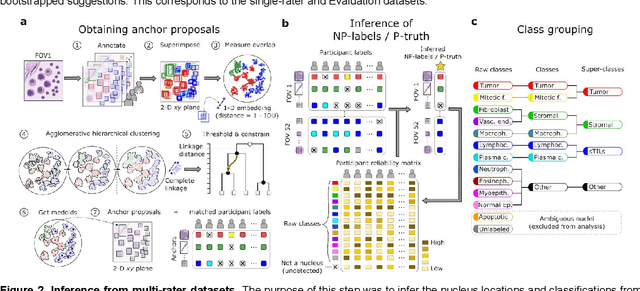
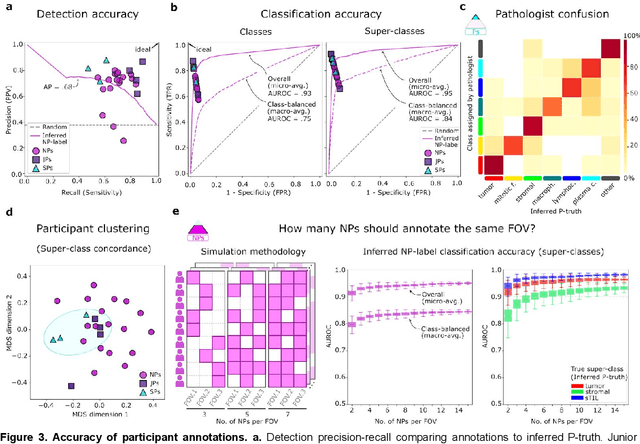
Abstract:High-resolution mapping of cells and tissue structures provides a foundation for developing interpretable machine-learning models for computational pathology. Deep learning algorithms can provide accurate mappings given large numbers of labeled instances for training and validation. Generating adequate volume of quality labels has emerged as a critical barrier in computational pathology given the time and effort required from pathologists. In this paper we describe an approach for engaging crowds of medical students and pathologists that was used to produce a dataset of over 220,000 annotations of cell nuclei in breast cancers. We show how suggested annotations generated by a weak algorithm can improve the accuracy of annotations generated by non-experts and can yield useful data for training segmentation algorithms without laborious manual tracing. We systematically examine interrater agreement and describe modifications to the MaskRCNN model to improve cell mapping. We also describe a technique we call Decision Tree Approximation of Learned Embeddings (DTALE) that leverages nucleus segmentations and morphologic features to improve the transparency of nucleus classification models. The annotation data produced in this study are freely available for algorithm development and benchmarking at: https://sites.google.com/view/nucls.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge