Amina Mollaysa
GRAM-TDI: adaptive multimodal representation learning for drug target interaction prediction
Sep 26, 2025Abstract:Drug target interaction (DTI) prediction is a cornerstone of computational drug discovery, enabling rational design, repurposing, and mechanistic insights. While deep learning has advanced DTI modeling, existing approaches primarily rely on SMILES protein pairs and fail to exploit the rich multimodal information available for small molecules and proteins. We introduce GRAMDTI, a pretraining framework that integrates multimodal molecular and protein inputs into unified representations. GRAMDTI extends volume based contrastive learning to four modalities, capturing higher-order semantic alignment beyond conventional pairwise approaches. To handle modality informativeness, we propose adaptive modality dropout, dynamically regulating each modality's contribution during pre-training. Additionally, IC50 activity measurements, when available, are incorporated as weak supervision to ground representations in biologically meaningful interaction strengths. Experiments on four publicly available datasets demonstrate that GRAMDTI consistently outperforms state of the art baselines. Our results highlight the benefits of higher order multimodal alignment, adaptive modality utilization, and auxiliary supervision for robust and generalizable DTI prediction.
TRIDENT: Tri-Modal Molecular Representation Learning with Taxonomic Annotations and Local Correspondence
Jun 26, 2025Abstract:Molecular property prediction aims to learn representations that map chemical structures to functional properties. While multimodal learning has emerged as a powerful paradigm to learn molecular representations, prior works have largely overlooked textual and taxonomic information of molecules for representation learning. We introduce TRIDENT, a novel framework that integrates molecular SMILES, textual descriptions, and taxonomic functional annotations to learn rich molecular representations. To achieve this, we curate a comprehensive dataset of molecule-text pairs with structured, multi-level functional annotations. Instead of relying on conventional contrastive loss, TRIDENT employs a volume-based alignment objective to jointly align tri-modal features at the global level, enabling soft, geometry-aware alignment across modalities. Additionally, TRIDENT introduces a novel local alignment objective that captures detailed relationships between molecular substructures and their corresponding sub-textual descriptions. A momentum-based mechanism dynamically balances global and local alignment, enabling the model to learn both broad functional semantics and fine-grained structure-function mappings. TRIDENT achieves state-of-the-art performance on 11 downstream tasks, demonstrating the value of combining SMILES, textual, and taxonomic functional annotations for molecular property prediction.
BioLangFusion: Multimodal Fusion of DNA, mRNA, and Protein Language Models
Jun 10, 2025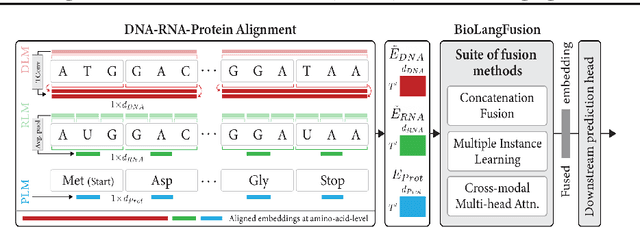
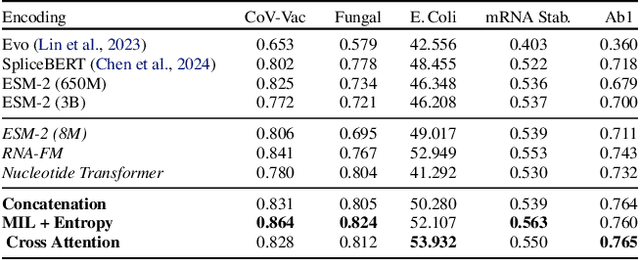
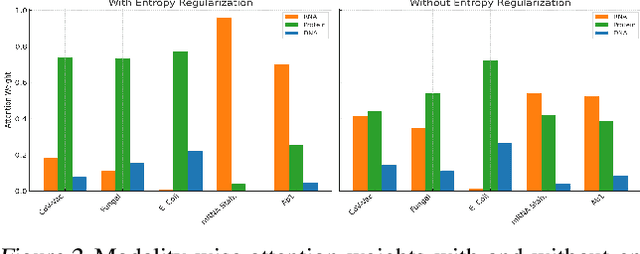
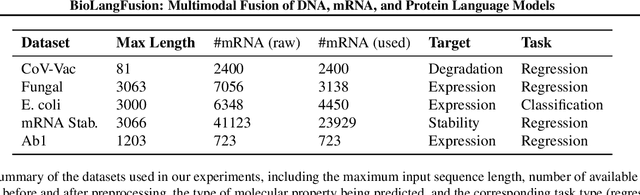
Abstract:We present BioLangFusion, a simple approach for integrating pre-trained DNA, mRNA, and protein language models into unified molecular representations. Motivated by the central dogma of molecular biology (information flow from gene to transcript to protein), we align per-modality embeddings at the biologically meaningful codon level (three nucleotides encoding one amino acid) to ensure direct cross-modal correspondence. BioLangFusion studies three standard fusion techniques: (i) codon-level embedding concatenation, (ii) entropy-regularized attention pooling inspired by multiple-instance learning, and (iii) cross-modal multi-head attention -- each technique providing a different inductive bias for combining modality-specific signals. These methods require no additional pre-training or modification of the base models, allowing straightforward integration with existing sequence-based foundation models. Across five molecular property prediction tasks, BioLangFusion outperforms strong unimodal baselines, showing that even simple fusion of pre-trained models can capture complementary multi-omic information with minimal overhead.
Attention-based Multi-task Learning for Base Editor Outcome Prediction
Nov 15, 2023



Abstract:Human genetic diseases often arise from point mutations, emphasizing the critical need for precise genome editing techniques. Among these, base editing stands out as it allows targeted alterations at the single nucleotide level. However, its clinical application is hindered by low editing efficiency and unintended mutations, necessitating extensive trial-and-error experimentation in the laboratory. To speed up this process, we present an attention-based two-stage machine learning model that learns to predict the likelihood of all possible editing outcomes for a given genomic target sequence. We further propose a multi-task learning schema to jointly learn multiple base editors (i.e. variants) at once. Our model's predictions consistently demonstrated a strong correlation with the actual experimental results on multiple datasets and base editor variants. These results provide further validation for the models' capacity to enhance and accelerate the process of refining base editing designs.
Dynamic Local Attention with Hierarchical Patching for Irregular Clinical Time Series
Nov 13, 2023



Abstract:Irregular multivariate time series data is prevalent in the clinical and healthcare domains. It is characterized by time-wise and feature-wise irregularities, making it challenging for machine learning methods to work with. To solve this, we introduce a new model architecture composed of two modules: (1) DLA, a Dynamic Local Attention mechanism that uses learnable queries and feature-specific local windows when computing the self-attention operation. This results in aggregating irregular time steps raw input within each window to a harmonized regular latent space representation while taking into account the different features' sampling rates. (2) A hierarchical MLP mixer that processes the output of DLA through multi-scale patching to leverage information at various scales for the downstream tasks. Our approach outperforms state-of-the-art methods on three real-world datasets, including the latest clinical MIMIC IV dataset.
Generating Personalized Insulin Treatments Strategies with Deep Conditional Generative Time Series Models
Sep 28, 2023



Abstract:We propose a novel framework that combines deep generative time series models with decision theory for generating personalized treatment strategies. It leverages historical patient trajectory data to jointly learn the generation of realistic personalized treatment and future outcome trajectories through deep generative time series models. In particular, our framework enables the generation of novel multivariate treatment strategies tailored to the personalized patient history and trained for optimal expected future outcomes based on conditional expected utility maximization. We demonstrate our framework by generating personalized insulin treatment strategies and blood glucose predictions for hospitalized diabetes patients, showcasing the potential of our approach for generating improved personalized treatment strategies. Keywords: deep generative model, probabilistic decision support, personalized treatment generation, insulin and blood glucose prediction
SimTS: Rethinking Contrastive Representation Learning for Time Series Forecasting
Mar 31, 2023



Abstract:Contrastive learning methods have shown an impressive ability to learn meaningful representations for image or time series classification. However, these methods are less effective for time series forecasting, as optimization of instance discrimination is not directly applicable to predicting the future state from the history context. Moreover, the construction of positive and negative pairs in current technologies strongly relies on specific time series characteristics, restricting their generalization across diverse types of time series data. To address these limitations, we propose SimTS, a simple representation learning approach for improving time series forecasting by learning to predict the future from the past in the latent space. SimTS does not rely on negative pairs or specific assumptions about the characteristics of the particular time series. Our extensive experiments on several benchmark time series forecasting datasets show that SimTS achieves competitive performance compared to existing contrastive learning methods. Furthermore, we show the shortcomings of the current contrastive learning framework used for time series forecasting through a detailed ablation study. Overall, our work suggests that SimTS is a promising alternative to other contrastive learning approaches for time series forecasting.
Goal-directed Generation of Discrete Structures with Conditional Generative Models
Oct 23, 2020



Abstract:Despite recent advances, goal-directed generation of structured discrete data remains challenging. For problems such as program synthesis (generating source code) and materials design (generating molecules), finding examples which satisfy desired constraints or exhibit desired properties is difficult. In practice, expensive heuristic search or reinforcement learning algorithms are often employed. In this paper we investigate the use of conditional generative models which directly attack this inverse problem, by modeling the distribution of discrete structures given properties of interest. Unfortunately, maximum likelihood training of such models often fails with the samples from the generative model inadequately respecting the input properties. To address this, we introduce a novel approach to directly optimize a reinforcement learning objective, maximizing an expected reward. We avoid high-variance score-function estimators that would otherwise be required by sampling from an approximation to the normalized rewards, allowing simple Monte Carlo estimation of model gradients. We test our methodology on two tasks: generating molecules with user-defined properties and identifying short python expressions which evaluate to a given target value. In both cases, we find improvements over maximum likelihood estimation and other baselines.
Regularising Non-linear Models Using Feature Side-information
Mar 07, 2017



Abstract:Very often features come with their own vectorial descriptions which provide detailed information about their properties. We refer to these vectorial descriptions as feature side-information. In the standard learning scenario, input is represented as a vector of features and the feature side-information is most often ignored or used only for feature selection prior to model fitting. We believe that feature side-information which carries information about features intrinsic property will help improve model prediction if used in a proper way during learning process. In this paper, we propose a framework that allows for the incorporation of the feature side-information during the learning of very general model families to improve the prediction performance. We control the structures of the learned models so that they reflect features similarities as these are defined on the basis of the side-information. We perform experiments on a number of benchmark datasets which show significant predictive performance gains, over a number of baselines, as a result of the exploitation of the side-information.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge