Junzhou Huang
GenePheno: Interpretable Gene Knockout-Induced Phenotype Abnormality Prediction from Gene Sequences
Nov 14, 2025
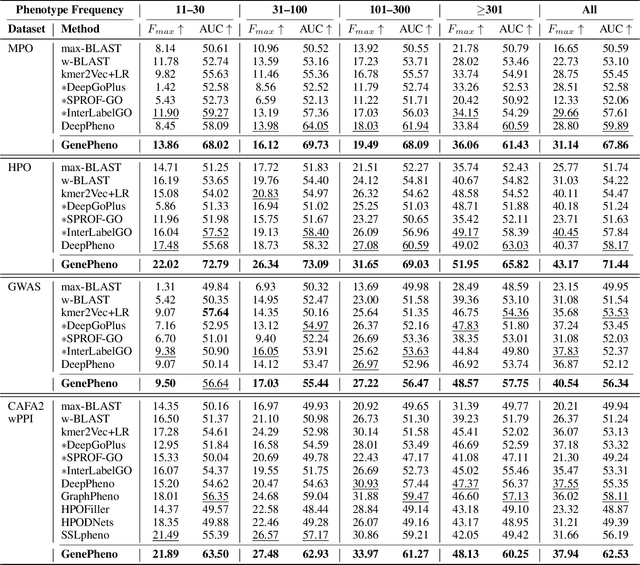
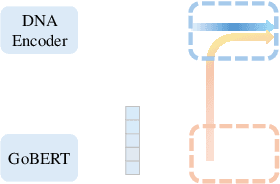
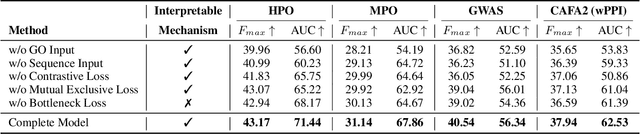
Abstract:Exploring how genetic sequences shape phenotypes is a fundamental challenge in biology and a key step toward scalable, hypothesis-driven experimentation. The task is complicated by the large modality gap between sequences and phenotypes, as well as the pleiotropic nature of gene-phenotype relationships. Existing sequence-based efforts focus on the degree to which variants of specific genes alter a limited set of phenotypes, while general gene knockout induced phenotype abnormality prediction methods heavily rely on curated genetic information as inputs, which limits scalability and generalizability. As a result, the task of broadly predicting the presence of multiple phenotype abnormalities under gene knockout directly from gene sequences remains underexplored. We introduce GenePheno, the first interpretable multi-label prediction framework that predicts knockout induced phenotypic abnormalities from gene sequences. GenePheno employs a contrastive multi-label learning objective that captures inter-phenotype correlations, complemented by an exclusive regularization that enforces biological consistency. It further incorporates a gene function bottleneck layer, offering human interpretable concepts that reflect functional mechanisms behind phenotype formation. To support progress in this area, we curate four datasets with canonical gene sequences as input and multi-label phenotypic abnormalities induced by gene knockouts as targets. Across these datasets, GenePheno achieves state-of-the-art gene-centric $F_{\text{max}}$ and phenotype-centric AUC, and case studies demonstrate its ability to reveal gene functional mechanisms.
GRAM-TDI: adaptive multimodal representation learning for drug target interaction prediction
Sep 26, 2025Abstract:Drug target interaction (DTI) prediction is a cornerstone of computational drug discovery, enabling rational design, repurposing, and mechanistic insights. While deep learning has advanced DTI modeling, existing approaches primarily rely on SMILES protein pairs and fail to exploit the rich multimodal information available for small molecules and proteins. We introduce GRAMDTI, a pretraining framework that integrates multimodal molecular and protein inputs into unified representations. GRAMDTI extends volume based contrastive learning to four modalities, capturing higher-order semantic alignment beyond conventional pairwise approaches. To handle modality informativeness, we propose adaptive modality dropout, dynamically regulating each modality's contribution during pre-training. Additionally, IC50 activity measurements, when available, are incorporated as weak supervision to ground representations in biologically meaningful interaction strengths. Experiments on four publicly available datasets demonstrate that GRAMDTI consistently outperforms state of the art baselines. Our results highlight the benefits of higher order multimodal alignment, adaptive modality utilization, and auxiliary supervision for robust and generalizable DTI prediction.
TRIDENT: Tri-Modal Molecular Representation Learning with Taxonomic Annotations and Local Correspondence
Jun 26, 2025Abstract:Molecular property prediction aims to learn representations that map chemical structures to functional properties. While multimodal learning has emerged as a powerful paradigm to learn molecular representations, prior works have largely overlooked textual and taxonomic information of molecules for representation learning. We introduce TRIDENT, a novel framework that integrates molecular SMILES, textual descriptions, and taxonomic functional annotations to learn rich molecular representations. To achieve this, we curate a comprehensive dataset of molecule-text pairs with structured, multi-level functional annotations. Instead of relying on conventional contrastive loss, TRIDENT employs a volume-based alignment objective to jointly align tri-modal features at the global level, enabling soft, geometry-aware alignment across modalities. Additionally, TRIDENT introduces a novel local alignment objective that captures detailed relationships between molecular substructures and their corresponding sub-textual descriptions. A momentum-based mechanism dynamically balances global and local alignment, enabling the model to learn both broad functional semantics and fine-grained structure-function mappings. TRIDENT achieves state-of-the-art performance on 11 downstream tasks, demonstrating the value of combining SMILES, textual, and taxonomic functional annotations for molecular property prediction.
AI-Assisted Decision-Making for Clinical Assessment of Auto-Segmented Contour Quality
May 01, 2025Abstract:Purpose: This study presents a Deep Learning (DL)-based quality assessment (QA) approach for evaluating auto-generated contours (auto-contours) in radiotherapy, with emphasis on Online Adaptive Radiotherapy (OART). Leveraging Bayesian Ordinal Classification (BOC) and calibrated uncertainty thresholds, the method enables confident QA predictions without relying on ground truth contours or extensive manual labeling. Methods: We developed a BOC model to classify auto-contour quality and quantify prediction uncertainty. A calibration step was used to optimize uncertainty thresholds that meet clinical accuracy needs. The method was validated under three data scenarios: no manual labels, limited labels, and extensive labels. For rectum contours in prostate cancer, we applied geometric surrogate labels when manual labels were absent, transfer learning when limited, and direct supervision when ample labels were available. Results: The BOC model delivered robust performance across all scenarios. Fine-tuning with just 30 manual labels and calibrating with 34 subjects yielded over 90% accuracy on test data. Using the calibrated threshold, over 93% of the auto-contours' qualities were accurately predicted in over 98% of cases, reducing unnecessary manual reviews and highlighting cases needing correction. Conclusion: The proposed QA model enhances contouring efficiency in OART by reducing manual workload and enabling fast, informed clinical decisions. Through uncertainty quantification, it ensures safer, more reliable radiotherapy workflows.
Leveraging Gait Patterns as Biomarkers: An attention-guided Deep Multiple Instance Learning Network for Scoliosis Classification
Apr 04, 2025Abstract:Scoliosis is a spinal curvature disorder that is difficult to detect early and can compress the chest cavity, impacting respiratory function and cardiac health. Especially for adolescents, delayed detection and treatment result in worsening compression. Traditional scoliosis detection methods heavily rely on clinical expertise, and X-ray imaging poses radiation risks, limiting large-scale early screening. We propose an Attention-Guided Deep Multi-Instance Learning method (Gait-MIL) to effectively capture discriminative features from gait patterns, which is inspired by ScoNet-MT's pioneering use of gait patterns for scoliosis detection. We evaluate our method on the first large-scale dataset based on gait patterns for scoliosis classification. The results demonstrate that our study improves the performance of using gait as a biomarker for scoliosis detection, significantly enhances detection accuracy for the particularly challenging Neutral cases, where subtle indicators are often overlooked. Our Gait-MIL also performs robustly in imbalanced scenarios, making it a promising tool for large-scale scoliosis screening.
InversionGNN: A Dual Path Network for Multi-Property Molecular Optimization
Mar 03, 2025



Abstract:Exploring chemical space to find novel molecules that simultaneously satisfy multiple properties is crucial in drug discovery. However, existing methods often struggle with trading off multiple properties due to the conflicting or correlated nature of chemical properties. To tackle this issue, we introduce InversionGNN framework, an effective yet sample-efficient dual-path graph neural network (GNN) for multi-objective drug discovery. In the direct prediction path of InversionGNN, we train the model for multi-property prediction to acquire knowledge of the optimal combination of functional groups. Then the learned chemical knowledge helps the inversion generation path to generate molecules with required properties. In order to decode the complex knowledge of multiple properties in the inversion path, we propose a gradient-based Pareto search method to balance conflicting properties and generate Pareto optimal molecules. Additionally, InversionGNN is able to search the full Pareto front approximately in discrete chemical space. Comprehensive experimental evaluations show that InversionGNN is both effective and sample-efficient in various discrete multi-objective settings including drug discovery.
MLLM4PUE: Toward Universal Embeddings in Computational Pathology through Multimodal LLMs
Feb 11, 2025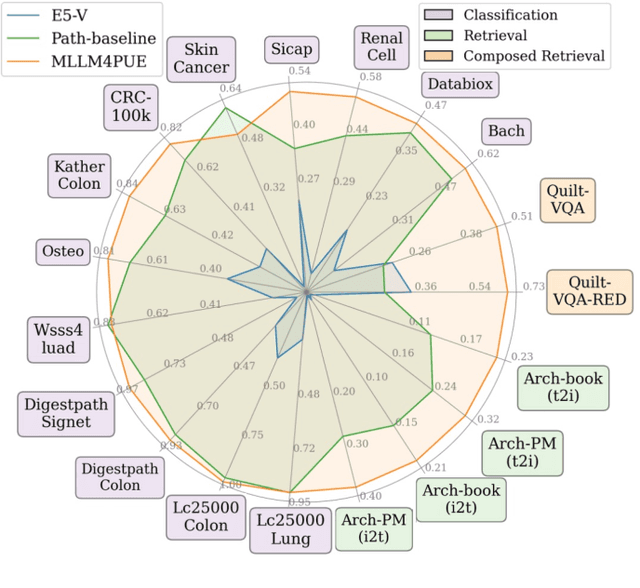
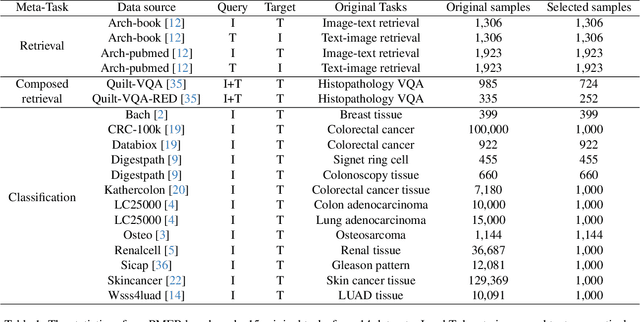
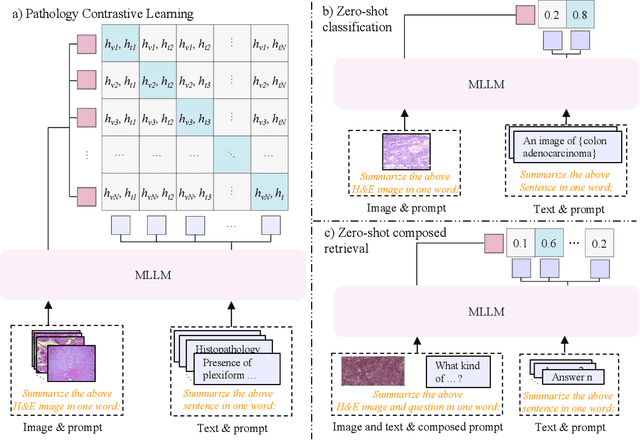
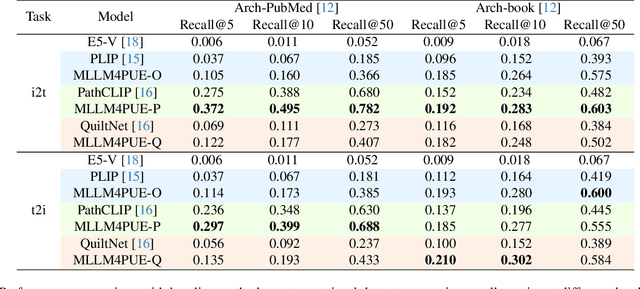
Abstract:Pathology plays a critical role in diagnosing a wide range of diseases, yet existing approaches often rely heavily on task-specific models trained on extensive, well-labeled datasets. These methods face sustainability challenges due to the diversity of pathologies and the labor-intensive nature of data collection. To address these limitations, we highlight the need for universal multimodal embeddings that can support multiple downstream tasks. Previous approaches often involve fine-tuning CLIP-based models, which handle images and text separately, limiting their ability to capture complex multimodal relationships. Additionally, these models are evaluated across diverse datasets without a unified benchmark for assessing multimodal embeddings in pathology. To address these challenges, we propose MLLM4PUE, a novel framework that leverages Multimodal Large Language Models (MLLMs) to generate Pathology Universal Embeddings. The MLLM4PUE framework not only facilitates robust integration of images and text but also enhances understanding and fusion capabilities across various tasks. We further introduce the Pathology Multimodal Embedding Benchmark (PMEB), a comprehensive benchmark designed to assess the quality of pathology multimodal embeddings. PMEB comprises 15 original tasks drawn from 14 datasets, organized into three meta-tasks: retrieval, classification, and composed retrieval. Experimental results demonstrate the superiority of MLLM4PUE, illustrating MLLM-based models can effectively support a wide range of downstream tasks and unify the research direction for foundation models in pathology.
Natural Language-Assisted Multi-modal Medication Recommendation
Jan 13, 2025Abstract:Combinatorial medication recommendation(CMR) is a fundamental task of healthcare, which offers opportunities for clinical physicians to provide more precise prescriptions for patients with intricate health conditions, particularly in the scenarios of long-term medical care. Previous research efforts have sought to extract meaningful information from electronic health records (EHRs) to facilitate combinatorial medication recommendations. Existing learning-based approaches further consider the chemical structures of medications, but ignore the textual medication descriptions in which the functionalities are clearly described. Furthermore, the textual knowledge derived from the EHRs of patients remains largely underutilized. To address these issues, we introduce the Natural Language-Assisted Multi-modal Medication Recommendation(NLA-MMR), a multi-modal alignment framework designed to learn knowledge from the patient view and medication view jointly. Specifically, NLA-MMR formulates CMR as an alignment problem from patient and medication modalities. In this vein, we employ pretrained language models(PLMs) to extract in-domain knowledge regarding patients and medications, serving as the foundational representation for both modalities. In the medication modality, we exploit both chemical structures and textual descriptions to create medication representations. In the patient modality, we generate the patient representations based on textual descriptions of diagnosis, procedure, and symptom. Extensive experiments conducted on three publicly accessible datasets demonstrate that NLA-MMR achieves new state-of-the-art performance, with a notable average improvement of 4.72% in Jaccard score. Our source code is publicly available on https://github.com/jtan1102/NLA-MMR_CIKM_2024.
* 10 pages
GoBERT: Gene Ontology Graph Informed BERT for Universal Gene Function Prediction
Jan 03, 2025Abstract:Exploring the functions of genes and gene products is crucial to a wide range of fields, including medical research, evolutionary biology, and environmental science. However, discovering new functions largely relies on expensive and exhaustive wet lab experiments. Existing methods of automatic function annotation or prediction mainly focus on protein function prediction with sequence, 3D-structures or protein family information. In this study, we propose to tackle the gene function prediction problem by exploring Gene Ontology graph and annotation with BERT (GoBERT) to decipher the underlying relationships among gene functions. Our proposed novel function prediction task utilizes existing functions as inputs and generalizes the function prediction to gene and gene products. Specifically, two pre-train tasks are designed to jointly train GoBERT to capture both explicit and implicit relations of functions. Neighborhood prediction is a self-supervised multi-label classification task that captures the explicit function relations. Specified masking and recovering task helps GoBERT in finding implicit patterns among functions. The pre-trained GoBERT possess the ability to predict novel functions for various gene and gene products based on known functional annotations. Extensive experiments, biological case studies, and ablation studies are conducted to demonstrate the superiority of our proposed GoBERT.
Compositional Image Retrieval via Instruction-Aware Contrastive Learning
Dec 07, 2024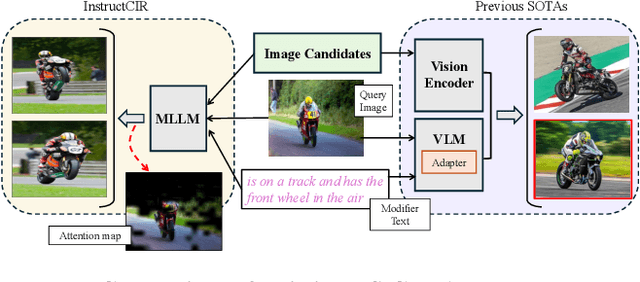
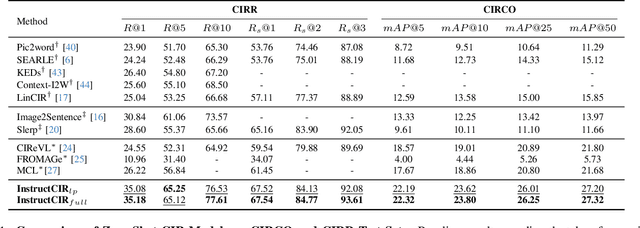
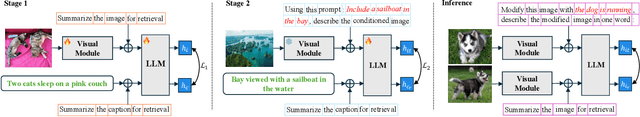
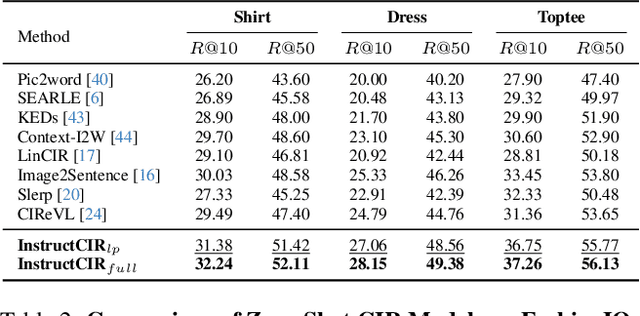
Abstract:Composed Image Retrieval (CIR) involves retrieving a target image based on a composed query of an image paired with text that specifies modifications or changes to the visual reference. CIR is inherently an instruction-following task, as the model needs to interpret and apply modifications to the image. In practice, due to the scarcity of annotated data in downstream tasks, Zero-Shot CIR (ZS-CIR) is desirable. While existing ZS-CIR models based on CLIP have shown promising results, their capability in interpreting and following modification instructions remains limited. Some research attempts to address this by incorporating Large Language Models (LLMs). However, these approaches still face challenges in effectively integrating multimodal information and instruction understanding. To tackle above challenges, we propose a novel embedding method utilizing an instruction-tuned Multimodal LLM (MLLM) to generate composed representation, which significantly enhance the instruction following capability for a comprehensive integration between images and instructions. Nevertheless, directly applying MLLMs introduces a new challenge since MLLMs are primarily designed for text generation rather than embedding extraction as required in CIR. To address this, we introduce a two-stage training strategy to efficiently learn a joint multimodal embedding space and further refining the ability to follow modification instructions by tuning the model in a triplet dataset similar to the CIR format. Extensive experiments on four public datasets: FashionIQ, CIRR, GeneCIS, and CIRCO demonstrates the superior performance of our model, outperforming state-of-the-art baselines by a significant margin. Codes are available at the GitHub repository.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge