Ti Bai
Medical Artificial Intelligence and Automation Laboratory and Department of Radiation Oncology, UT Southwestern Medical Center, Dallas TX 75235, USA
AI-Assisted Decision-Making for Clinical Assessment of Auto-Segmented Contour Quality
May 01, 2025Abstract:Purpose: This study presents a Deep Learning (DL)-based quality assessment (QA) approach for evaluating auto-generated contours (auto-contours) in radiotherapy, with emphasis on Online Adaptive Radiotherapy (OART). Leveraging Bayesian Ordinal Classification (BOC) and calibrated uncertainty thresholds, the method enables confident QA predictions without relying on ground truth contours or extensive manual labeling. Methods: We developed a BOC model to classify auto-contour quality and quantify prediction uncertainty. A calibration step was used to optimize uncertainty thresholds that meet clinical accuracy needs. The method was validated under three data scenarios: no manual labels, limited labels, and extensive labels. For rectum contours in prostate cancer, we applied geometric surrogate labels when manual labels were absent, transfer learning when limited, and direct supervision when ample labels were available. Results: The BOC model delivered robust performance across all scenarios. Fine-tuning with just 30 manual labels and calibrating with 34 subjects yielded over 90% accuracy on test data. Using the calibrated threshold, over 93% of the auto-contours' qualities were accurately predicted in over 98% of cases, reducing unnecessary manual reviews and highlighting cases needing correction. Conclusion: The proposed QA model enhances contouring efficiency in OART by reducing manual workload and enabling fast, informed clinical decisions. Through uncertainty quantification, it ensures safer, more reliable radiotherapy workflows.
Deep Learning (DL)-based Automatic Segmentation of the Internal Pudendal Artery (IPA) for Reduction of Erectile Dysfunction in Definitive Radiotherapy of Localized Prostate Cancer
Feb 03, 2023Abstract:Background and purpose: Radiation-induced erectile dysfunction (RiED) is commonly seen in prostate cancer patients. Clinical trials have been developed in multiple institutions to investigate whether dose-sparing to the internal-pudendal-arteries (IPA) will improve retention of sexual potency. The IPA is usually not considered a conventional organ-at-risk (OAR) due to segmentation difficulty. In this work, we propose a deep learning (DL)-based auto-segmentation model for the IPA that utilizes CT and MRI or CT alone as the input image modality to accommodate variation in clinical practice. Materials and methods: 86 patients with CT and MRI images and noisy IPA labels were recruited in this study. We split the data into 42/14/30 for model training, testing, and a clinical observer study, respectively. There were three major innovations in this model: 1) we designed an architecture with squeeze-and-excite blocks and modality attention for effective feature extraction and production of accurate segmentation, 2) a novel loss function was used for training the model effectively with noisy labels, and 3) modality dropout strategy was used for making the model capable of segmentation in the absence of MRI. Results: The DSC, ASD, and HD95 values for the test dataset were 62.2%, 2.54mm, and 7mm, respectively. AI segmented contours were dosimetrically equivalent to the expert physician's contours. The observer study showed that expert physicians' scored AI contours (mean=3.7) higher than inexperienced physicians' contours (mean=3.1). When inexperienced physicians started with AI contours, the score improved to 3.7. Conclusion: The proposed model achieved good quality IPA contours to improve uniformity of segmentation and to facilitate introduction of standardized IPA segmentation into clinical trials and practice.
Coarse-Super-Resolution-Fine Network (CoSF-Net): A Unified End-to-End Neural Network for 4D-MRI with Simultaneous Motion Estimation and Super-Resolution
Nov 21, 2022



Abstract:Four-dimensional magnetic resonance imaging (4D-MRI) is an emerging technique for tumor motion management in image-guided radiation therapy (IGRT). However, current 4D-MRI suffers from low spatial resolution and strong motion artifacts owing to the long acquisition time and patients' respiratory variations; these limitations, if not managed properly, can adversely affect treatment planning and delivery in IGRT. Herein, we developed a novel deep learning framework called the coarse-super-resolution-fine network (CoSF-Net) to achieve simultaneous motion estimation and super-resolution in a unified model. We designed CoSF-Net by fully excavating the inherent properties of 4D-MRI, with consideration of limited and imperfectly matched training datasets. We conducted extensive experiments on multiple real patient datasets to verify the feasibility and robustness of the developed network. Compared with existing networks and three state-of-the-art conventional algorithms, CoSF-Net not only accurately estimated the deformable vector fields between the respiratory phases of 4D-MRI but also simultaneously improved the spatial resolution of 4D-MRI with enhanced anatomic features, yielding 4D-MR images with high spatiotemporal resolution.
Prior Guided Deep Difference Meta-Learner for Fast Adaptation to Stylized Segmentation
Nov 19, 2022Abstract:When a pre-trained general auto-segmentation model is deployed at a new institution, a support framework in the proposed Prior-guided DDL network will learn the systematic difference between the model predictions and the final contours revised and approved by clinicians for an initial group of patients. The learned style feature differences are concatenated with the new patients (query) features and then decoded to get the style-adapted segmentations. The model is independent of practice styles and anatomical structures. It meta-learns with simulated style differences and does not need to be exposed to any real clinical stylized structures during training. Once trained on the simulated data, it can be deployed for clinical use to adapt to new practice styles and new anatomical structures without further training. To show the proof of concept, we tested the Prior-guided DDL network on six different practice style variations for three different anatomical structures. Pre-trained segmentation models were adapted from post-operative clinical target volume (CTV) segmentation to segment CTVstyle1, CTVstyle2, and CTVstyle3, from parotid gland segmentation to segment Parotidsuperficial, and from rectum segmentation to segment Rectumsuperior and Rectumposterior. The mode performance was quantified with Dice Similarity Coefficient (DSC). With adaptation based on only the first three patients, the average DSCs were improved from 78.6, 71.9, 63.0, 52.2, 46.3 and 69.6 to 84.4, 77.8, 73.0, 77.8, 70.5, 68.1, for CTVstyle1, CTVstyle2, and CTVstyle3, Parotidsuperficial, Rectumsuperior, and Rectumposterior, respectively, showing the great potential of the Priorguided DDL network for a fast and effortless adaptation to new practice styles
Performance Deterioration of Deep Learning Models after Clinical Deployment: A Case Study with Auto-segmentation for Definitive Prostate Cancer Radiotherapy
Oct 11, 2022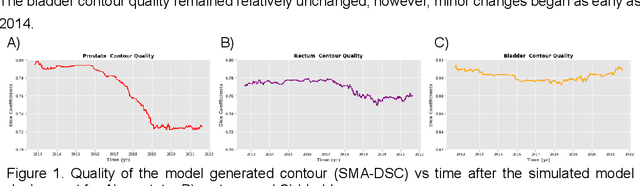
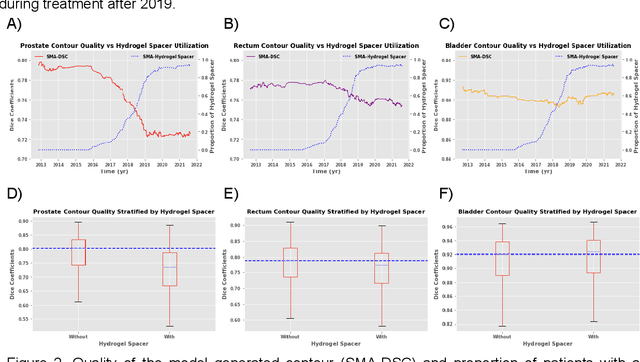
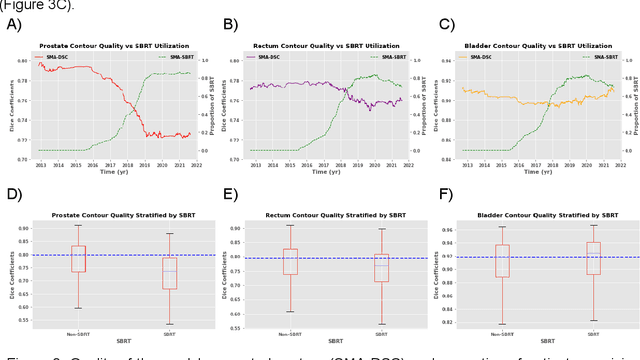
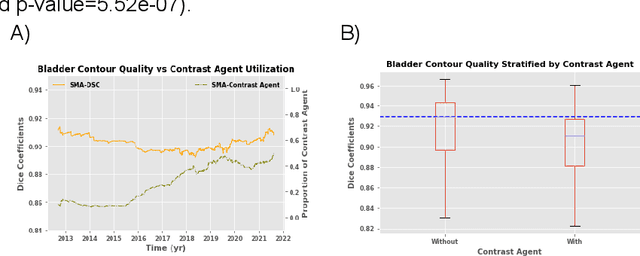
Abstract:In the past decade, deep learning (DL)-based artificial intelligence (AI) has witnessed unprecedented success and has led to much excitement in medicine. However, many successful models have not been implemented in the clinic predominantly due to concerns regarding the lack of interpretability and generalizability in both spatial and temporal domains. In this work, we used a DL-based auto segmentation model for intact prostate patients to observe any temporal performance changes and then correlate them to possible explanatory variables. We retrospectively simulated the clinical implementation of our DL model to investigate temporal performance trends. Our cohort included 912 patients with prostate cancer treated with definitive radiotherapy from January 2006 to August 2021 at the University of Texas Southwestern Medical Center (UTSW). We trained a U-Net-based DL auto segmentation model on the data collected before 2012 and tested it on data collected from 2012 to 2021 to simulate the clinical deployment of the trained model starting in 2012. We visualize the trends using a simple moving average curve and used ANOVA and t-test to investigate the impact of various clinical factors. The prostate and rectum contour quality decreased rapidly after 2016-2017. Stereotactic body radiotherapy (SBRT) and hydrogel spacer use were significantly associated with prostate contour quality (p=5.6e-12 and 0.002, respectively). SBRT and physicians' styles are significantly associated with the rectum contour quality (p=0.0005 and 0.02, respectively). Only the presence of contrast within the bladder significantly affected the bladder contour quality (p=1.6e-7). We showed that DL model performance decreased over time in concordance with changes in clinical practice patterns and changes in clinical personnel.
Uncertainty estimations methods for a deep learning model to aid in clinical decision-making -- a clinician's perspective
Oct 02, 2022



Abstract:Prediction uncertainty estimation has clinical significance as it can potentially quantify prediction reliability. Clinicians may trust 'blackbox' models more if robust reliability information is available, which may lead to more models being adopted into clinical practice. There are several deep learning-inspired uncertainty estimation techniques, but few are implemented on medical datasets -- fewer on single institutional datasets/models. We sought to compare dropout variational inference (DO), test-time augmentation (TTA), conformal predictions, and single deterministic methods for estimating uncertainty using our model trained to predict feeding tube placement for 271 head and neck cancer patients treated with radiation. We compared the area under the curve (AUC), sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) trends for each method at various cutoffs that sought to stratify patients into 'certain' and 'uncertain' cohorts. These cutoffs were obtained by calculating the percentile "uncertainty" within the validation cohort and applied to the testing cohort. Broadly, the AUC, sensitivity, and NPV increased as the predictions were more 'certain' -- i.e., lower uncertainty estimates. However, when a majority vote (implementing 2/3 criteria: DO, TTA, conformal predictions) or a stricter approach (3/3 criteria) were used, AUC, sensitivity, and NPV improved without a notable loss in specificity or PPV. Especially for smaller, single institutional datasets, it may be important to evaluate multiple estimations techniques before incorporating a model into clinical practice.
Exploring the combination of deep-learning based direct segmentation and deformable image registration for cone-beam CT based auto-segmentation for adaptive radiotherapy
Jun 07, 2022



Abstract:CBCT-based online adaptive radiotherapy (ART) calls for accurate auto-segmentation models to reduce the time cost for physicians to edit contours, since the patient is immobilized on the treatment table waiting for treatment to start. However, auto-segmentation of CBCT images is a difficult task, majorly due to low image quality and lack of true labels for training a deep learning (DL) model. Meanwhile CBCT auto-segmentation in ART is a unique task compared to other segmentation problems, where manual contours on planning CT (pCT) are available. To make use of this prior knowledge, we propose to combine deformable image registration (DIR) and direct segmentation (DS) on CBCT for head and neck patients. First, we use deformed pCT contours derived from multiple DIR methods between pCT and CBCT as pseudo labels for training. Second, we use deformed pCT contours as bounding box to constrain the region of interest for DS. Meanwhile deformed pCT contours are used as pseudo labels for training, but are generated from different DIR algorithms from bounding box. Third, we fine-tune the model with bounding box on true labels. We found that DS on CBCT trained with pseudo labels and without utilizing any prior knowledge has very poor segmentation performance compared to DIR-only segmentation. However, adding deformed pCT contours as bounding box in the DS network can dramatically improve segmentation performance, comparable to DIR-only segmentation. The DS model with bounding box can be further improved by fine-tuning it with some real labels. Experiments showed that 7 out of 19 structures have at least 0.2 dice similarity coefficient increase compared to DIR-only segmentation. Utilizing deformed pCT contours as pseudo labels for training and as bounding box for shape and location feature extraction in a DS model is a good way to combine DIR and DS.
Region Specific Optimization (RSO)-based Deep Interactive Registration
Mar 08, 2022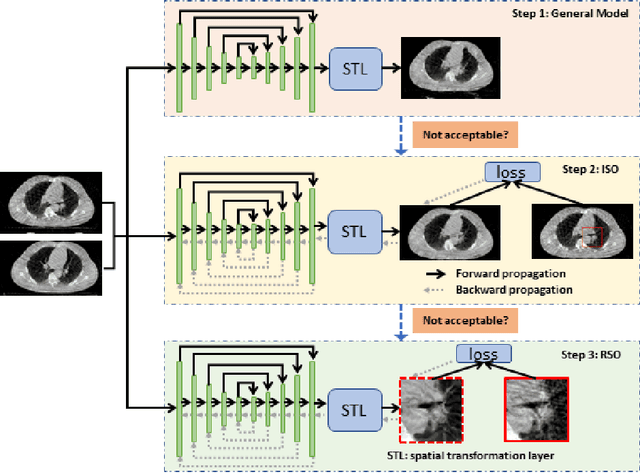
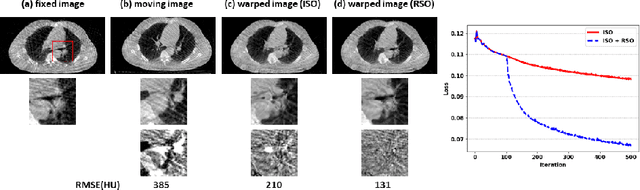

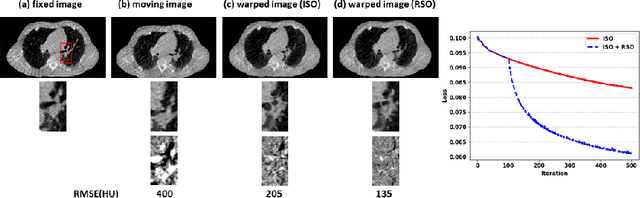
Abstract:Medical image registration is a fundamental and vital task which will affect the efficacy of many downstream clinical tasks. Deep learning (DL)-based deformable image registration (DIR) methods have been investigated, showing state-of-the-art performance. A test time optimization (TTO) technique was proposed to further improve the DL models' performance. Despite the substantial accuracy improvement with this TTO technique, there still remained some regions that exhibited large registration errors even after many TTO iterations. To mitigate this challenge, we firstly identified the reason why the TTO technique was slow, or even failed, to improve those regions' registration results. We then proposed a two-levels TTO technique, i.e., image-specific optimization (ISO) and region-specific optimization (RSO), where the region can be interactively indicated by the clinician during the registration result reviewing process. For both efficiency and accuracy, we further envisioned a three-step DL-based image registration workflow. Experimental results showed that our proposed method outperformed the conventional method qualitatively and quantitatively.
Segmentation by Test-Time Optimization (TTO) for CBCT-based Adaptive Radiation Therapy
Feb 08, 2022



Abstract:Online adaptive radiotherapy (ART) requires accurate and efficient auto-segmentation of target volumes and organs-at-risk (OARs) in mostly cone-beam computed tomography (CBCT) images. Propagating expert-drawn contours from the pre-treatment planning CT (pCT) through traditional or deep learning (DL) based deformable image registration (DIR) can achieve improved results in many situations. Typical DL-based DIR models are population based, that is, trained with a dataset for a population of patients, so they may be affected by the generalizability problem. In this paper, we propose a method called test-time optimization (TTO) to refine a pre-trained DL-based DIR population model, first for each individual test patient, and then progressively for each fraction of online ART treatment. Our proposed method is less susceptible to the generalizability problem, and thus can improve overall performance of different DL-based DIR models by improving model accuracy, especially for outliers. Our experiments used data from 239 patients with head and neck squamous cell carcinoma to test the proposed method. Firstly, we trained a population model with 200 patients, and then applied TTO to the remaining 39 test patients by refining the trained population model to obtain 39 individualized models. We compared each of the individualized models with the population model in terms of segmentation accuracy. The number of patients with at least 0.05 DSC improvement or 2 mm HD95 improvement by TTO averaged over the 17 selected structures for the state-of-the-art architecture Voxelmorph is 10 out of 39 test patients. The average time for deriving the individualized model using TTO from the pre-trained population model is approximately four minutes. When adapting the individualized model to a later fraction of the same patient, the average time is reduced to about one minute and the accuracy is slightly improved.
S2MS: Self-Supervised Learning Driven Multi-Spectral CT Image Enhancement
Jan 26, 2022



Abstract:Photon counting spectral CT (PCCT) can produce reconstructed attenuation maps in different energy channels, reflecting energy properties of the scanned object. Due to the limited photon numbers and the non-ideal detector response of each energy channel, the reconstructed images usually contain much noise. With the development of Deep Learning (DL) technique, different kinds of DL-based models have been proposed for noise reduction. However, most of the models require clean data set as the training labels, which are not always available in medical imaging field. Inspiring by the similarities of each channel's reconstructed image, we proposed a self-supervised learning based PCCT image enhancement framework via multi-spectral channels (S2MS). In S2MS framework, both the input and output labels are noisy images. Specifically, one single channel image was used as output while images of other single channels and channel-sum image were used as input to train the network, which can fully use the spectral data information without extra cost. The simulation results based on the AAPM Low-dose CT Challenge database showed that the proposed S2MS model can suppress the noise and preserve details more effectively in comparison with the traditional DL models, which has potential to improve the image quality of PCCT in clinical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge