Anjali Balagopal
Deep Learning (DL)-based Automatic Segmentation of the Internal Pudendal Artery (IPA) for Reduction of Erectile Dysfunction in Definitive Radiotherapy of Localized Prostate Cancer
Feb 03, 2023Abstract:Background and purpose: Radiation-induced erectile dysfunction (RiED) is commonly seen in prostate cancer patients. Clinical trials have been developed in multiple institutions to investigate whether dose-sparing to the internal-pudendal-arteries (IPA) will improve retention of sexual potency. The IPA is usually not considered a conventional organ-at-risk (OAR) due to segmentation difficulty. In this work, we propose a deep learning (DL)-based auto-segmentation model for the IPA that utilizes CT and MRI or CT alone as the input image modality to accommodate variation in clinical practice. Materials and methods: 86 patients with CT and MRI images and noisy IPA labels were recruited in this study. We split the data into 42/14/30 for model training, testing, and a clinical observer study, respectively. There were three major innovations in this model: 1) we designed an architecture with squeeze-and-excite blocks and modality attention for effective feature extraction and production of accurate segmentation, 2) a novel loss function was used for training the model effectively with noisy labels, and 3) modality dropout strategy was used for making the model capable of segmentation in the absence of MRI. Results: The DSC, ASD, and HD95 values for the test dataset were 62.2%, 2.54mm, and 7mm, respectively. AI segmented contours were dosimetrically equivalent to the expert physician's contours. The observer study showed that expert physicians' scored AI contours (mean=3.7) higher than inexperienced physicians' contours (mean=3.1). When inexperienced physicians started with AI contours, the score improved to 3.7. Conclusion: The proposed model achieved good quality IPA contours to improve uniformity of segmentation and to facilitate introduction of standardized IPA segmentation into clinical trials and practice.
Prior Guided Deep Difference Meta-Learner for Fast Adaptation to Stylized Segmentation
Nov 19, 2022Abstract:When a pre-trained general auto-segmentation model is deployed at a new institution, a support framework in the proposed Prior-guided DDL network will learn the systematic difference between the model predictions and the final contours revised and approved by clinicians for an initial group of patients. The learned style feature differences are concatenated with the new patients (query) features and then decoded to get the style-adapted segmentations. The model is independent of practice styles and anatomical structures. It meta-learns with simulated style differences and does not need to be exposed to any real clinical stylized structures during training. Once trained on the simulated data, it can be deployed for clinical use to adapt to new practice styles and new anatomical structures without further training. To show the proof of concept, we tested the Prior-guided DDL network on six different practice style variations for three different anatomical structures. Pre-trained segmentation models were adapted from post-operative clinical target volume (CTV) segmentation to segment CTVstyle1, CTVstyle2, and CTVstyle3, from parotid gland segmentation to segment Parotidsuperficial, and from rectum segmentation to segment Rectumsuperior and Rectumposterior. The mode performance was quantified with Dice Similarity Coefficient (DSC). With adaptation based on only the first three patients, the average DSCs were improved from 78.6, 71.9, 63.0, 52.2, 46.3 and 69.6 to 84.4, 77.8, 73.0, 77.8, 70.5, 68.1, for CTVstyle1, CTVstyle2, and CTVstyle3, Parotidsuperficial, Rectumsuperior, and Rectumposterior, respectively, showing the great potential of the Priorguided DDL network for a fast and effortless adaptation to new practice styles
A Proof-of-Concept Study of Artificial Intelligence Assisted Contour Revision
Jul 28, 2021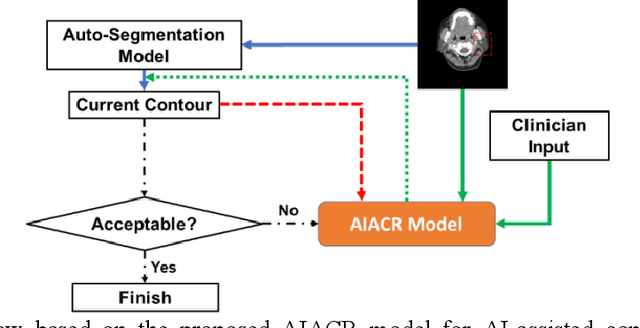

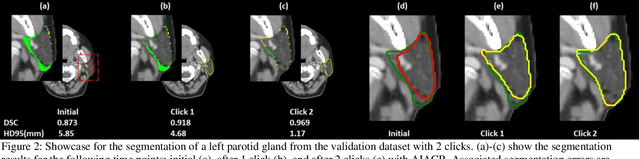
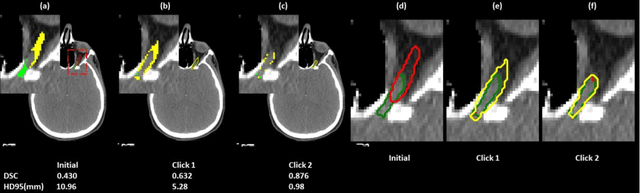
Abstract:Automatic segmentation of anatomical structures is critical for many medical applications. However, the results are not always clinically acceptable and require tedious manual revision. Here, we present a novel concept called artificial intelligence assisted contour revision (AIACR) and demonstrate its feasibility. The proposed clinical workflow of AIACR is as follows given an initial contour that requires a clinicians revision, the clinician indicates where a large revision is needed, and a trained deep learning (DL) model takes this input to update the contour. This process repeats until a clinically acceptable contour is achieved. The DL model is designed to minimize the clinicians input at each iteration and to minimize the number of iterations needed to reach acceptance. In this proof-of-concept study, we demonstrated the concept on 2D axial images of three head-and-neck cancer datasets, with the clinicians input at each iteration being one mouse click on the desired location of the contour segment. The performance of the model is quantified with Dice Similarity Coefficient (DSC) and 95th percentile of Hausdorff Distance (HD95). The average DSC/HD95 (mm) of the auto-generated initial contours were 0.82/4.3, 0.73/5.6 and 0.67/11.4 for three datasets, which were improved to 0.91/2.1, 0.86/2.4 and 0.86/4.7 with three mouse clicks, respectively. Each DL-based contour update requires around 20 ms. We proposed a novel AIACR concept that uses DL models to assist clinicians in revising contours in an efficient and effective way, and we demonstrated its feasibility by using 2D axial CT images from three head-and-neck cancer datasets.
Site-Agnostic 3D Dose Distribution Prediction with Deep Learning Neural Networks
Jun 15, 2021
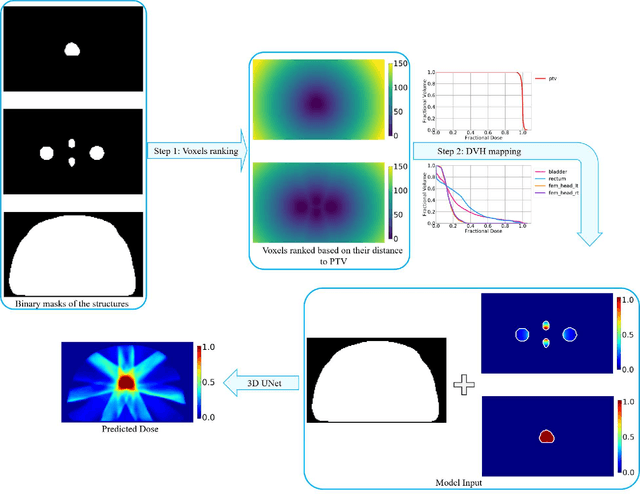
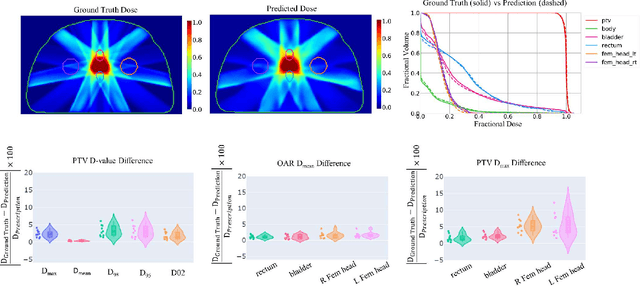
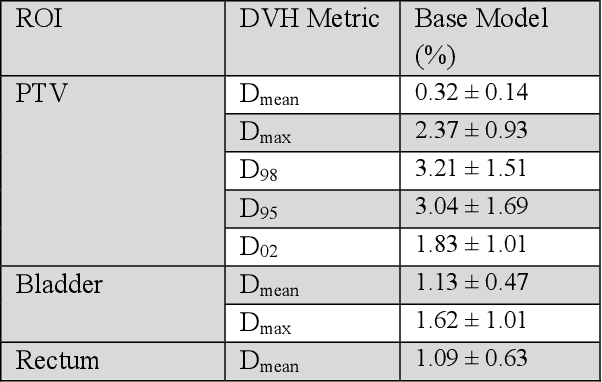
Abstract:Typically, the current dose prediction models are limited to small amounts of data and require re-training for a specific site, often leading to suboptimal performance. We propose a site-agnostic, 3D dose distribution prediction model using deep learning that can leverage data from any treatment site, thus increasing the total data available to train the model. Applying our proposed model to a new target treatment site requires only a brief fine-tuning of the model to the new data and involves no modifications to the model input channels or its parameters. Thus, it can be efficiently adapted to a different treatment site, even with a small training dataset.
PSA-Net: Deep Learning based Physician Style-Aware Segmentation Network for Post-Operative Prostate Cancer Clinical Target Volume
Feb 15, 2021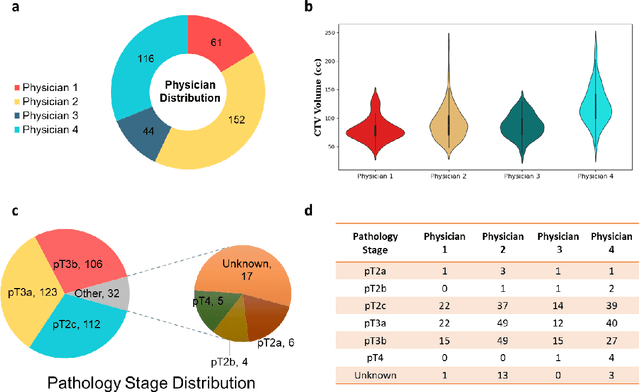
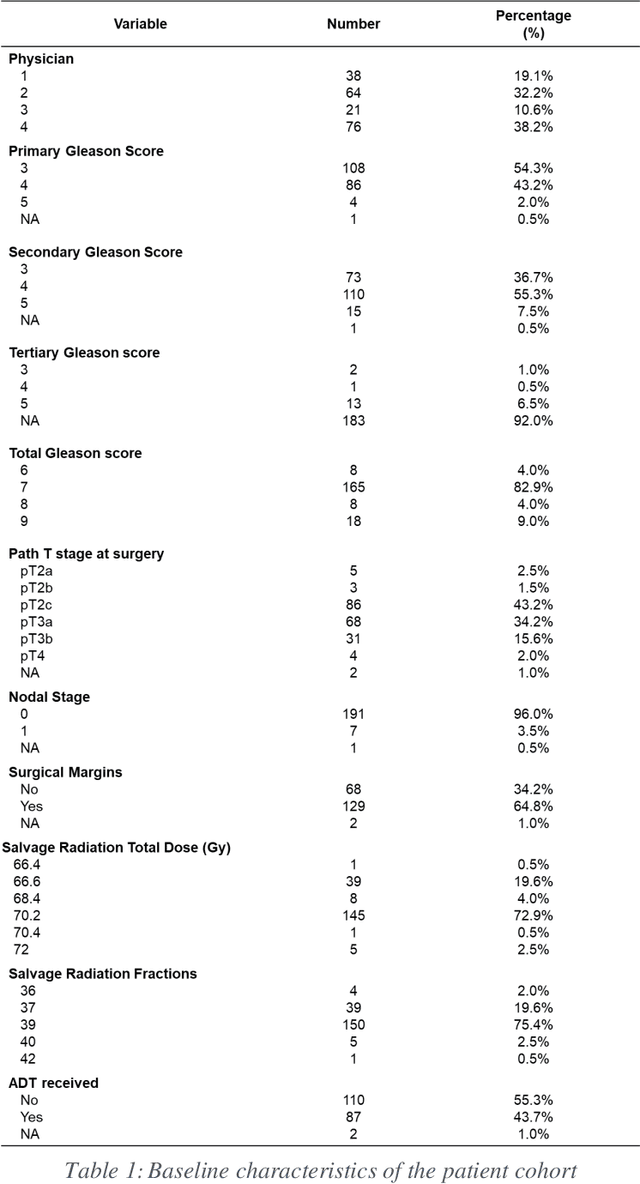
Abstract:Automatic segmentation of medical images with DL algorithms has proven to be highly successful. With most of these algorithms, inter-observer variation is an acknowledged problem, leading to sub-optimal results. This problem is even more significant in post-operative clinical target volume (post-op CTV) segmentation due to the absence of macroscopic visual tumor in the image. This study, using post-op CTV segmentation as the test bed, tries to determine if physician styles are consistent and learnable, if there is an impact of physician styles on treatment outcome and toxicity; and how to explicitly deal with physician styles in DL algorithms to facilitate its clinical acceptance. A classifier is trained to identify which physician has contoured the CTV from just the contour and corresponding CT scan, to determine if physician styles are consistent and learnable. Next, we evaluate if adapting automatic segmentation to physician styles would be clinically feasible based on a lack of difference between outcomes. For modeling different physician styles of CTV segmentation, a concept called physician style-aware (PSA) segmentation is proposed which is an encoder-multidecoder network trained with perceptual loss. With the proposed physician style-aware network (PSA-Net), Dice similarity coefficient (DSC) accuracy increases on an average of 3.4% for all physicians from a general model that is not style adapted. We show that stylistic contouring variations also exist between institutions that follow the same segmentation guidelines and show the effectiveness of the proposed method in adapting to new institutional styles. We observed an accuracy improvement of 5% in terms of DSC when adapting to the style of a separate institution.
Dosimetric impact of physician style variations in contouring CTV for post-operative prostate cancer: A deep learning based simulation study
Feb 01, 2021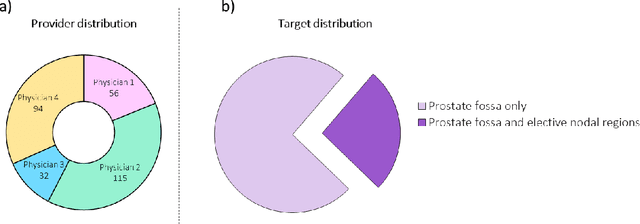
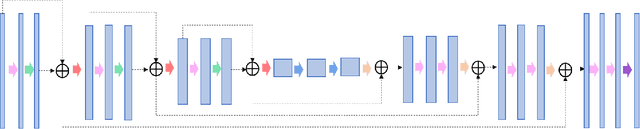
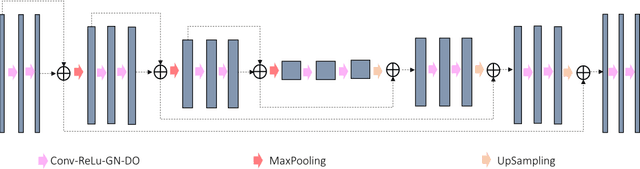
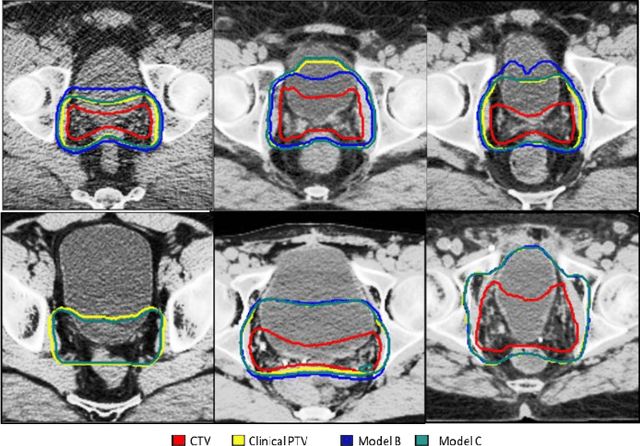
Abstract:In tumor segmentation, inter-observer variation is acknowledged to be a significant problem. This is even more significant in clinical target volume (CTV) segmentation, specifically, in post-operative settings, where a gross tumor does not exist. In this scenario, CTV is not an anatomically established structure but rather one determined by the physician based on the clinical guideline used, the preferred trade off between tumor control and toxicity, their experience, training background etc... This results in high inter-observer variability between physicians. Inter-observer variability has been considered an issue, however its dosimetric consequence is still unclear, due to the absence of multiple physician CTV contours for each patient and the significant amount of time required for dose planning. In this study, we analyze the impact that these physician stylistic variations have on organs-at-risk (OAR) dose by simulating the clinical workflow using deep learning. For a given patient previously treated by one physician, we use DL-based tools to simulate how other physicians would contour the CTV and how the corresponding dose distributions should look like for this patient. To simulate multiple physician styles, we use a previously developed in-house CTV segmentation model that can produce physician style-aware segmentations. The corresponding dose distribution is predicted using another in-house deep learning tool, which, averaging across all structures, is capable of predicting dose within 3% of the prescription dose on the test data. For every test patient, four different physician-style CTVs are considered and four different dose distributions are analyzed. OAR dose metrics are compared, showing that even though physician style variations results in organs getting different doses, all the important dose metrics except Maximum Dose point are within the clinically acceptable limit.
Using Monte Carlo dropout and bootstrap aggregation for uncertainty estimation in radiation therapy dose prediction with deep learning neural networks
Nov 01, 2020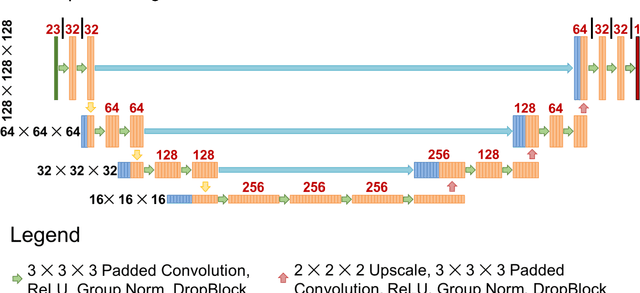
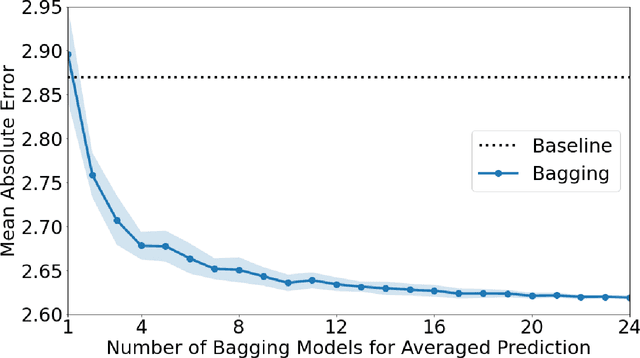
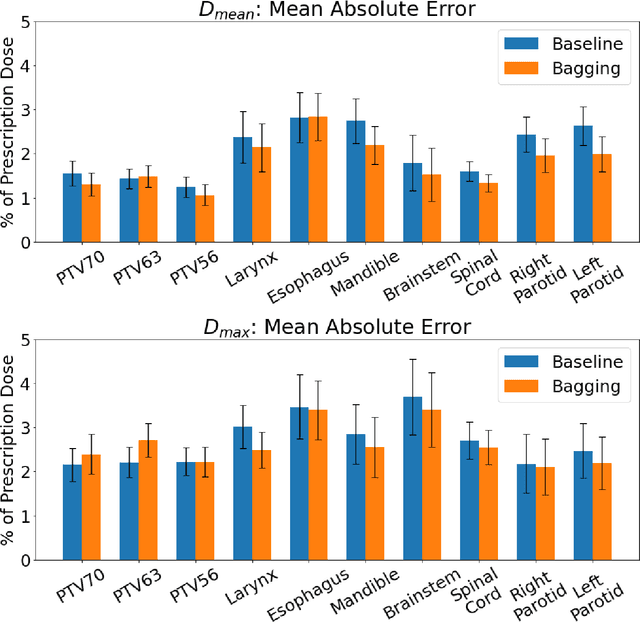
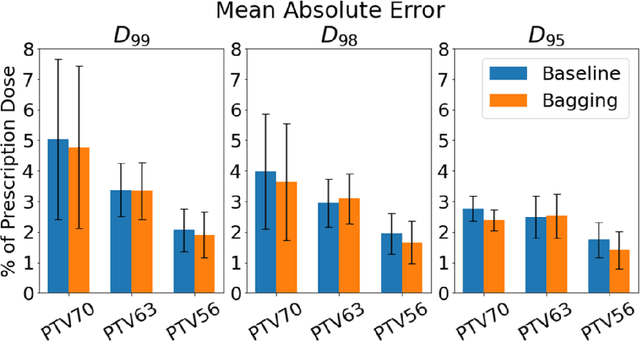
Abstract:Recently, artificial intelligence technologies and algorithms have become a major focus for advancements in treatment planning for radiation therapy. As these are starting to become incorporated into the clinical workflow, a major concern from clinicians is not whether the model is accurate, but whether the model can express to a human operator when it does not know if its answer is correct. We propose to use Monte Carlo dropout (MCDO) and the bootstrap aggregation (bagging) technique on deep learning models to produce uncertainty estimations for radiation therapy dose prediction. We show that both models are capable of generating a reasonable uncertainty map, and, with our proposed scaling technique, creating interpretable uncertainties and bounds on the prediction and any relevant metrics. Performance-wise, bagging provides statistically significant reduced loss value and errors in most of the metrics investigated in this study. The addition of bagging was able to further reduce errors by another 0.34% for Dmean and 0.19% for Dmax, on average, when compared to the baseline framework. Overall, the bagging framework provided significantly lower MAE of 2.62, as opposed to the baseline framework's MAE of 2.87. The usefulness of bagging, from solely a performance standpoint, does highly depend on the problem and the acceptable predictive error, and its high upfront computational cost during training should be factored in to deciding whether it is advantageous to use it. In terms of deployment with uncertainty estimations turned on, both frameworks offer the same performance time of about 12 seconds. As an ensemble-based metaheuristic, bagging can be used with existing machine learning architectures to improve stability and performance, and MCDO can be applied to any deep learning models that have dropout as part of their architecture.
A deep learning-based framework for segmenting invisible clinical target volumes with estimated uncertainties for post-operative prostate cancer radiotherapy
Apr 28, 2020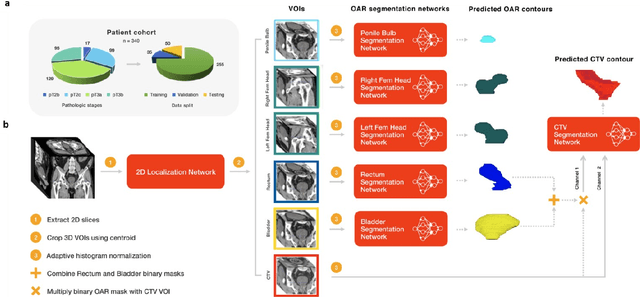
Abstract:In post-operative radiotherapy for prostate cancer, the cancerous prostate gland has been surgically removed, so the clinical target volume (CTV) to be irradiated encompasses the microscopic spread of tumor cells, which cannot be visualized in typical clinical images such as computed tomography or magnetic resonance imaging. In current clinical practice, physicians segment CTVs manually based on their relationship with nearby organs and other clinical information, per clinical guidelines. Automating post-operative prostate CTV segmentation with traditional image segmentation methods has been a major challenge. Here, we propose a deep learning model to overcome this problem by segmenting nearby organs first, then using their relationship with the CTV to assist CTV segmentation. The model proposed is trained using labels clinically approved and used for patient treatment, which are subject to relatively large inter-physician variations due to the absence of a visual ground truth. The model achieves an average Dice similarity coefficient (DSC) of 0.87 on a holdout dataset of 50 patients, much better than established methods, such as atlas-based methods (DSC<0.7). The uncertainties associated with automatically segmented CTV contours are also estimated to help physicians inspect and revise the contours, especially in areas with large inter-physician variations. We also use a 4-point grading system to show that the clinical quality of the automatically segmented CTV contours is equal to that of approved clinical contours manually drawn by physicians.
Fully Automated Organ Segmentation in Male Pelvic CT Images
May 31, 2018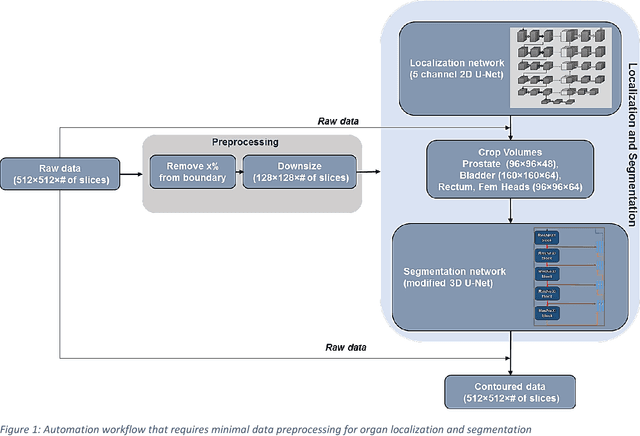
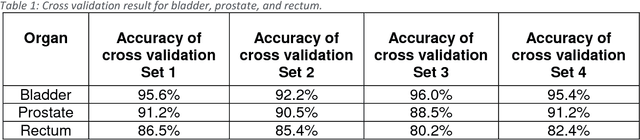
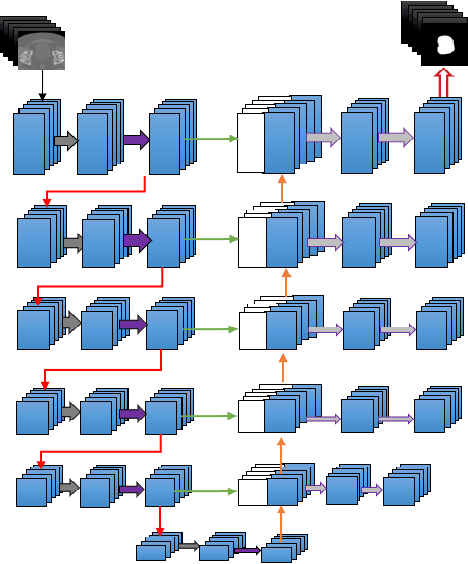
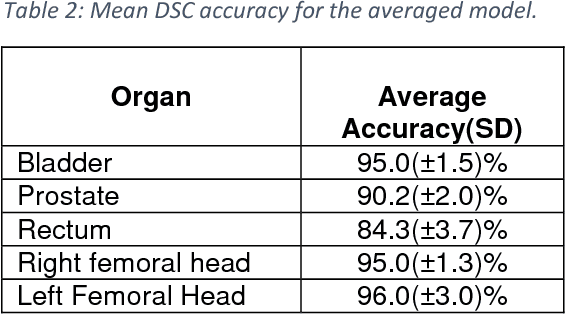
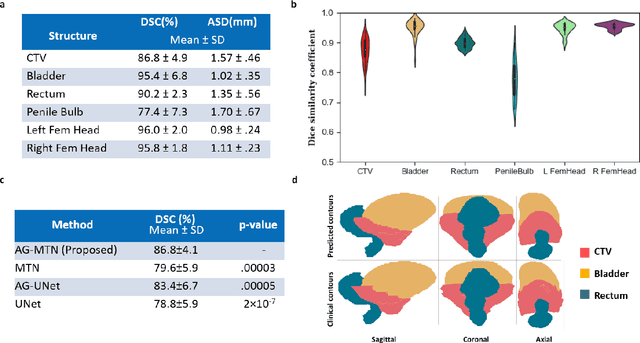
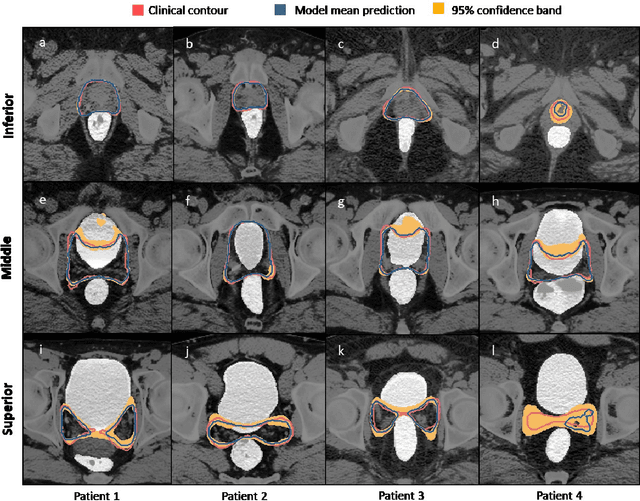
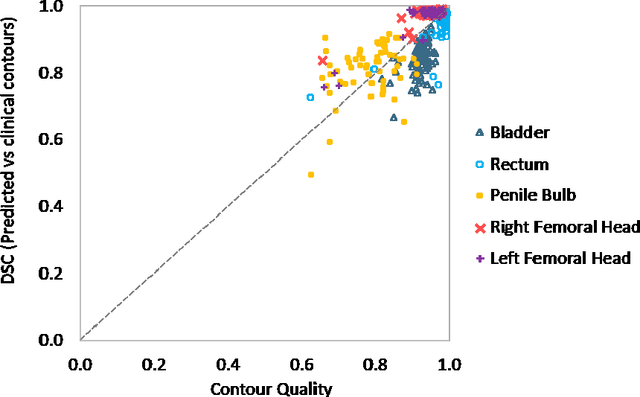
Abstract:Accurate segmentation of prostate and surrounding organs at risk is important for prostate cancer radiotherapy treatment planning. We present a fully automated workflow for male pelvic CT image segmentation using deep learning. The architecture consists of a 2D localization network followed by a 3D segmentation network for volumetric segmentation of prostate, bladder, rectum, and femoral heads. We used a multi-channel 2D U-Net followed by a 3D U-Net with encoding arm modified with aggregated residual networks, known as ResNeXt. The models were trained and tested on a pelvic CT image dataset comprising 136 patients. Test results show that 3D U-Net based segmentation achieves mean (SD) Dice coefficient values of 90 (2.0)% ,96 (3.0)%, 95 (1.3)%, 95 (1.5)%, and 84 (3.7)% for prostate, left femoral head, right femoral head, bladder, and rectum, respectively, using the proposed fully automated segmentation method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge