Azar Sadeghnejad Barkousaraie
Site-Agnostic 3D Dose Distribution Prediction with Deep Learning Neural Networks
Jun 15, 2021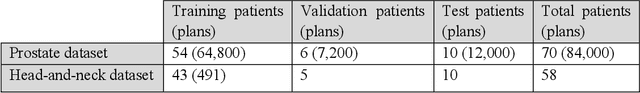
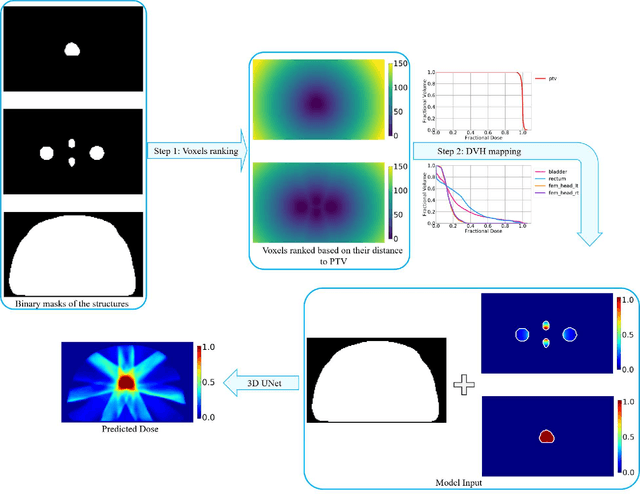
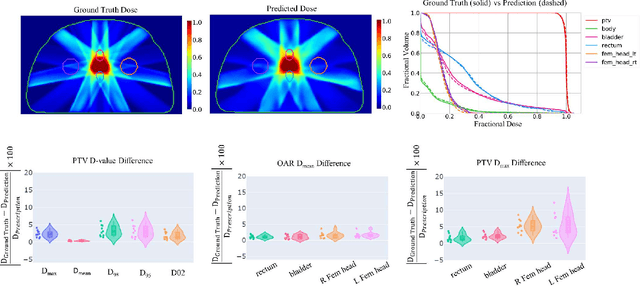
Abstract:Typically, the current dose prediction models are limited to small amounts of data and require re-training for a specific site, often leading to suboptimal performance. We propose a site-agnostic, 3D dose distribution prediction model using deep learning that can leverage data from any treatment site, thus increasing the total data available to train the model. Applying our proposed model to a new target treatment site requires only a brief fine-tuning of the model to the new data and involves no modifications to the model input channels or its parameters. Thus, it can be efficiently adapted to a different treatment site, even with a small training dataset.
Using Monte Carlo dropout and bootstrap aggregation for uncertainty estimation in radiation therapy dose prediction with deep learning neural networks
Nov 01, 2020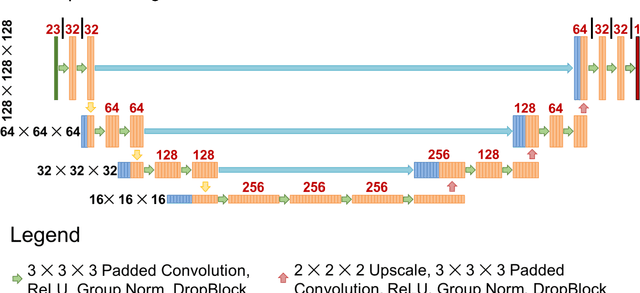
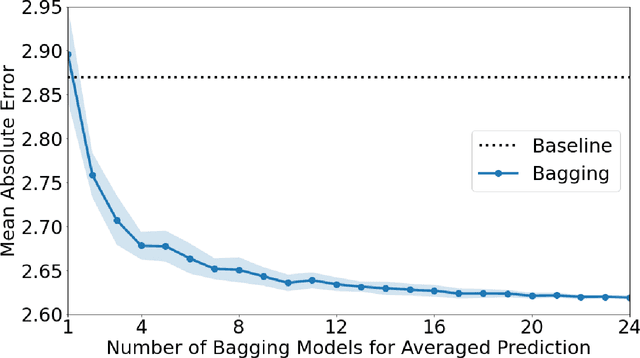
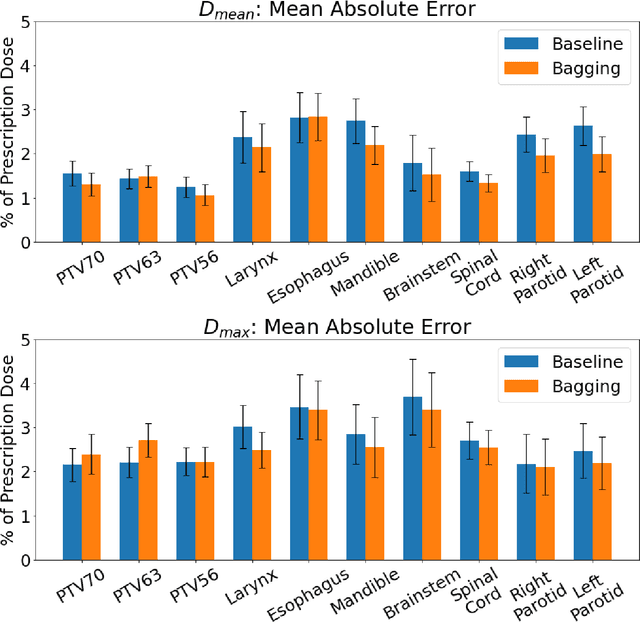
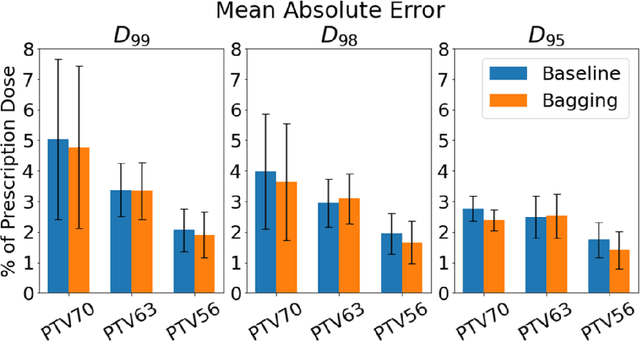
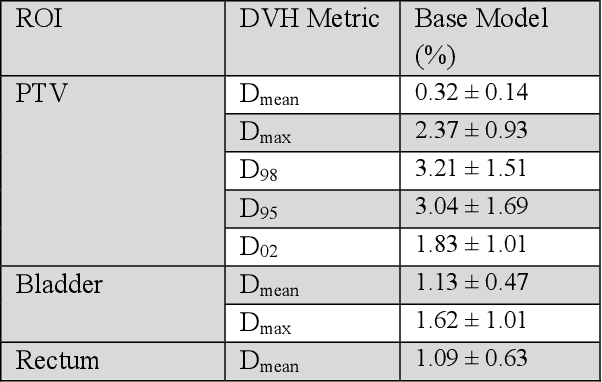
Abstract:Recently, artificial intelligence technologies and algorithms have become a major focus for advancements in treatment planning for radiation therapy. As these are starting to become incorporated into the clinical workflow, a major concern from clinicians is not whether the model is accurate, but whether the model can express to a human operator when it does not know if its answer is correct. We propose to use Monte Carlo dropout (MCDO) and the bootstrap aggregation (bagging) technique on deep learning models to produce uncertainty estimations for radiation therapy dose prediction. We show that both models are capable of generating a reasonable uncertainty map, and, with our proposed scaling technique, creating interpretable uncertainties and bounds on the prediction and any relevant metrics. Performance-wise, bagging provides statistically significant reduced loss value and errors in most of the metrics investigated in this study. The addition of bagging was able to further reduce errors by another 0.34% for Dmean and 0.19% for Dmax, on average, when compared to the baseline framework. Overall, the bagging framework provided significantly lower MAE of 2.62, as opposed to the baseline framework's MAE of 2.87. The usefulness of bagging, from solely a performance standpoint, does highly depend on the problem and the acceptable predictive error, and its high upfront computational cost during training should be factored in to deciding whether it is advantageous to use it. In terms of deployment with uncertainty estimations turned on, both frameworks offer the same performance time of about 12 seconds. As an ensemble-based metaheuristic, bagging can be used with existing machine learning architectures to improve stability and performance, and MCDO can be applied to any deep learning models that have dropout as part of their architecture.
Using Deep Learning to Predict Beam-Tunable Pareto Optimal Dose Distribution for Intensity Modulated Radiation Therapy
Jun 19, 2020

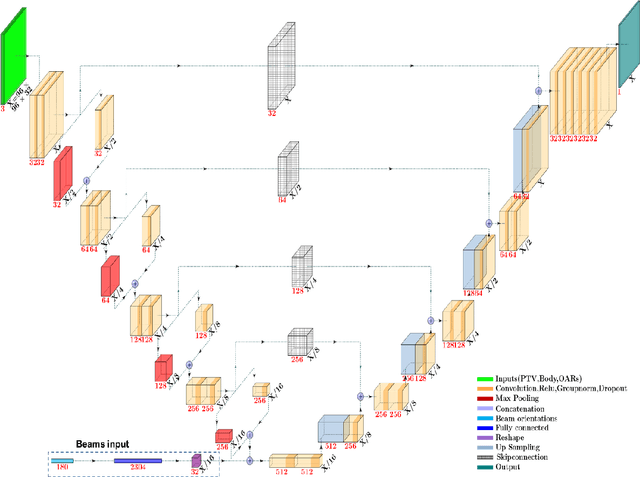
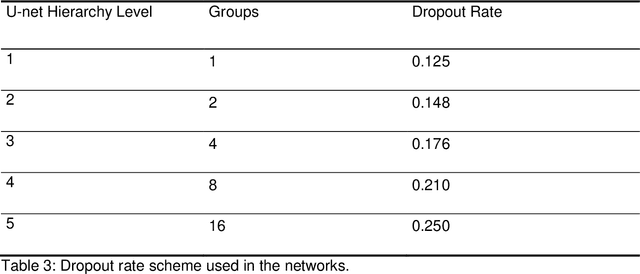
Abstract:We propose to develop deep learning models that can predict Pareto optimal dose distributions by using any given set of beam angles, along with patient anatomy, as input to train the deep neural networks. We implement and compare two deep learning networks that predict with two different beam configuration modalities. We generated Pareto optimal plans for 70 patients with prostate cancer. We used fluence map optimization to generate 500 IMRT plans that sampled the Pareto surface for each patient, for a total of 35,000 plans. We studied and compared two different models, Model I and Model II. Model I directly uses beam angles as a second input to the network as a binary vector. Model II converts the beam angles into beam doses that are conformal to the PTV. Our deep learning models predicted voxel-level dose distributions that precisely matched the ground truth dose distributions. Quantitatively, Model I prediction error of 0.043 (confirmation), 0.043 (homogeneity), 0.327 (R50), 2.80% (D95), 3.90% (D98), 0.6% (D50), 1.10% (D2) was lower than that of Model II, which obtained 0.076 (confirmation), 0.058 (homogeneity), 0.626 (R50), 7.10% (D95), 6.50% (D98), 8.40% (D50), 6.30% (D2). Treatment planners who use our models will be able to use deep learning to control the tradeoffs between the PTV and OAR weights, as well as the beam number and configurations in real time. Our dose prediction methods provide a stepping stone to building automatic IMRT treatment planning.
A deep learning-based framework for segmenting invisible clinical target volumes with estimated uncertainties for post-operative prostate cancer radiotherapy
Apr 28, 2020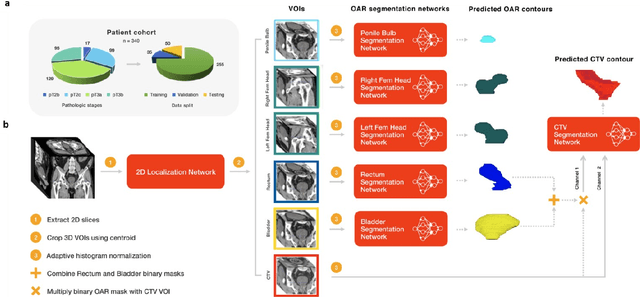
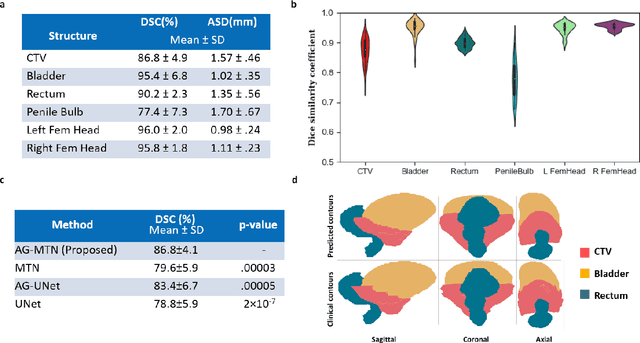
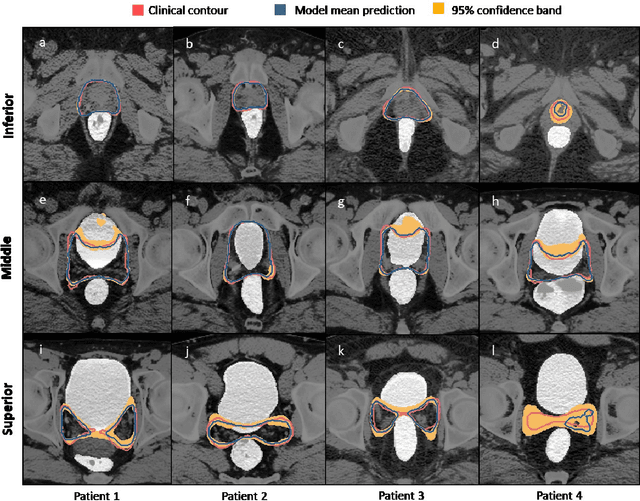
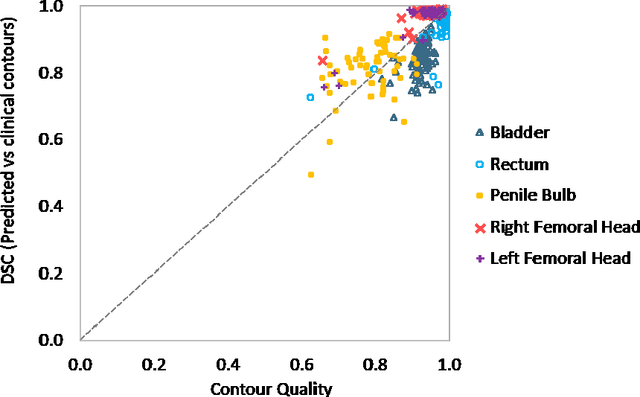
Abstract:In post-operative radiotherapy for prostate cancer, the cancerous prostate gland has been surgically removed, so the clinical target volume (CTV) to be irradiated encompasses the microscopic spread of tumor cells, which cannot be visualized in typical clinical images such as computed tomography or magnetic resonance imaging. In current clinical practice, physicians segment CTVs manually based on their relationship with nearby organs and other clinical information, per clinical guidelines. Automating post-operative prostate CTV segmentation with traditional image segmentation methods has been a major challenge. Here, we propose a deep learning model to overcome this problem by segmenting nearby organs first, then using their relationship with the CTV to assist CTV segmentation. The model proposed is trained using labels clinically approved and used for patient treatment, which are subject to relatively large inter-physician variations due to the absence of a visual ground truth. The model achieves an average Dice similarity coefficient (DSC) of 0.87 on a holdout dataset of 50 patients, much better than established methods, such as atlas-based methods (DSC<0.7). The uncertainties associated with automatically segmented CTV contours are also estimated to help physicians inspect and revise the contours, especially in areas with large inter-physician variations. We also use a 4-point grading system to show that the clinical quality of the automatically segmented CTV contours is equal to that of approved clinical contours manually drawn by physicians.
Incorporating human and learned domain knowledge into training deep neural networks: A differentiable dose volume histogram and adversarial inspired framework for generating Pareto optimal dose distributions in radiation therapy
Aug 16, 2019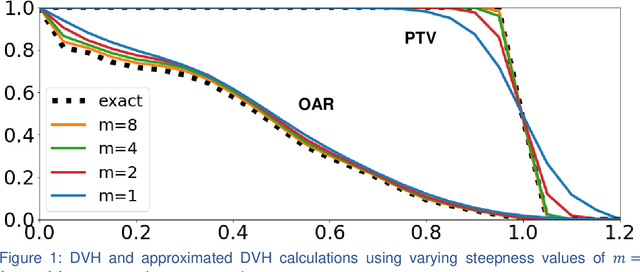
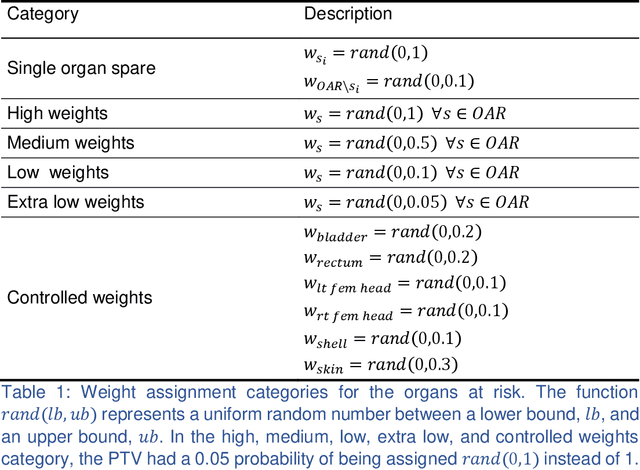
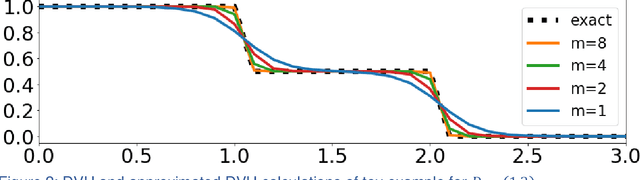

Abstract:We propose a novel domain specific loss, which is a differentiable loss function based on the dose volume histogram, and combine it with an adversarial loss for the training of deep neural networks to generate Pareto optimal dose distributions. The mean squared error (MSE) loss, dose volume histogram (DVH) loss, and adversarial (ADV) loss were used to train 4 instances of the neural network model: 1) MSE, 2) MSE+ADV, 3) MSE+DVH, and 4) MSE+DVH+ADV. 70 prostate patients were acquired, and the dose influence arrays were calculated for each patient. 1200 Pareto surface plans per patient were generated by pseudo-randomizing the tradeoff weights (84,000 plans total). We divided the data into 54 training, 6 validation, and 10 testing patients. Each model was trained for 100,000 iterations, with a batch size of 2. The prediction time of each model is 0.052 seconds. Quantitatively, the MSE+DVH+ADV model had the lowest prediction error of 0.038 (conformation), 0.026 (homogeneity), 0.298 (R50), 1.65% (D95), 2.14% (D98), 2.43% (D99). The MSE model had the worst prediction error of 0.134 (conformation), 0.041 (homogeneity), 0.520 (R50), 3.91% (D95), 4.33% (D98), 4.60% (D99). For both the mean dose PTV error and the max dose PTV, Body, Bladder and rectum error, the MSE+DVH+ADV outperformed all other models. All model's predictions have an average mean and max dose error less than 2.8% and 4.2%, respectively. Expert human domain specific knowledge can be the largest driver in the performance improvement, and adversarial learning can be used to further capture nuanced features. The real-time prediction capabilities allow for a physician to quickly navigate the tradeoff space, and produce a dose distribution as a tangible endpoint for the dosimetrist to use for planning. This can considerably reduce the treatment planning time, allowing for clinicians to focus their efforts on challenging cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge