Raquibul Hannan
A multimodal ensemble approach for clear cell renal cell carcinoma treatment outcome prediction
Dec 10, 2024



Abstract:Purpose: A reliable cancer prognosis model for clear cell renal cell carcinoma (ccRCC) can enhance personalized treatment. We developed a multi-modal ensemble model (MMEM) that integrates pretreatment clinical data, multi-omics data, and histopathology whole slide image (WSI) data to predict overall survival (OS) and disease-free survival (DFS) for ccRCC patients. Methods: We analyzed 226 patients from The Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma (TCGA-KIRC) dataset, which includes OS, DFS follow-up data, and five data modalities: clinical data, WSIs, and three multi-omics datasets (mRNA, miRNA, and DNA methylation). Separate survival models were built for OS and DFS. Cox-proportional hazards (CPH) model with forward feature selection is used for clinical and multi-omics data. Features from WSIs were extracted using ResNet and three general-purpose foundation models. A deep learning-based CPH model predicted survival using encoded WSI features. Risk scores from all models were combined based on training performance. Results: Performance was assessed using concordance index (C-index) and AUROC. The clinical feature-based CPH model received the highest weight for both OS and DFS tasks. Among WSI-based models, the general-purpose foundation model (UNI) achieved the best performance. The final MMEM model surpassed single-modality models, achieving C-indices of 0.820 (OS) and 0.833 (DFS), and AUROC values of 0.831 (3-year patient death) and 0.862 (cancer recurrence). Using predicted risk medians to stratify high- and low-risk groups, log-rank tests showed improved performance in both OS and DFS compared to single-modality models. Conclusion: MMEM is the first multi-modal model for ccRCC patients, integrating five data modalities. It outperformed single-modality models in prognostic ability and has the potential to assist in ccRCC patient management if independently validated.
Performance Deterioration of Deep Learning Models after Clinical Deployment: A Case Study with Auto-segmentation for Definitive Prostate Cancer Radiotherapy
Oct 11, 2022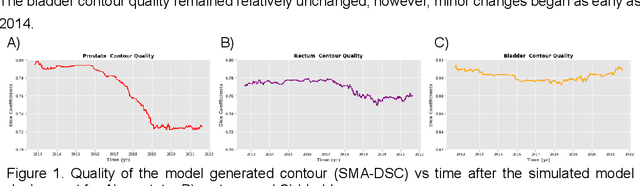
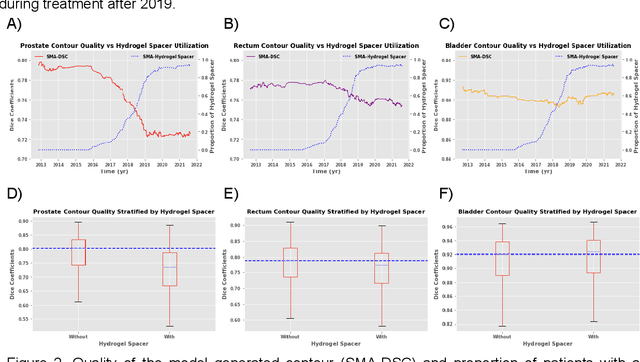
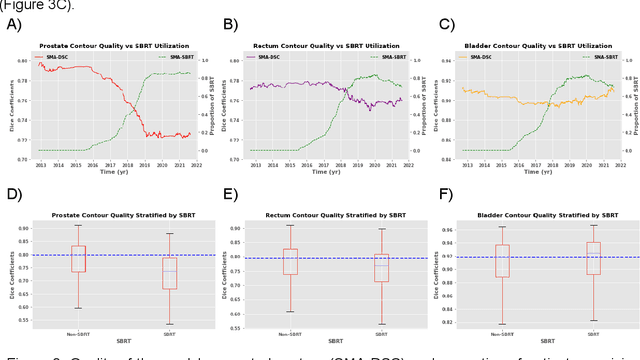
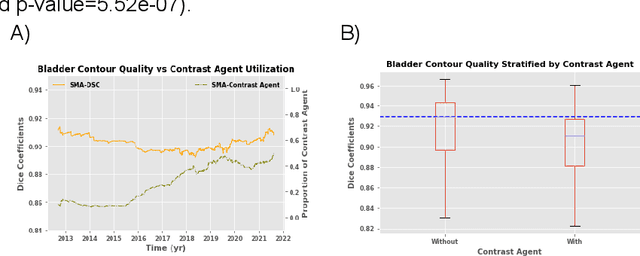
Abstract:In the past decade, deep learning (DL)-based artificial intelligence (AI) has witnessed unprecedented success and has led to much excitement in medicine. However, many successful models have not been implemented in the clinic predominantly due to concerns regarding the lack of interpretability and generalizability in both spatial and temporal domains. In this work, we used a DL-based auto segmentation model for intact prostate patients to observe any temporal performance changes and then correlate them to possible explanatory variables. We retrospectively simulated the clinical implementation of our DL model to investigate temporal performance trends. Our cohort included 912 patients with prostate cancer treated with definitive radiotherapy from January 2006 to August 2021 at the University of Texas Southwestern Medical Center (UTSW). We trained a U-Net-based DL auto segmentation model on the data collected before 2012 and tested it on data collected from 2012 to 2021 to simulate the clinical deployment of the trained model starting in 2012. We visualize the trends using a simple moving average curve and used ANOVA and t-test to investigate the impact of various clinical factors. The prostate and rectum contour quality decreased rapidly after 2016-2017. Stereotactic body radiotherapy (SBRT) and hydrogel spacer use were significantly associated with prostate contour quality (p=5.6e-12 and 0.002, respectively). SBRT and physicians' styles are significantly associated with the rectum contour quality (p=0.0005 and 0.02, respectively). Only the presence of contrast within the bladder significantly affected the bladder contour quality (p=1.6e-7). We showed that DL model performance decreased over time in concordance with changes in clinical practice patterns and changes in clinical personnel.
PSA-Net: Deep Learning based Physician Style-Aware Segmentation Network for Post-Operative Prostate Cancer Clinical Target Volume
Feb 15, 2021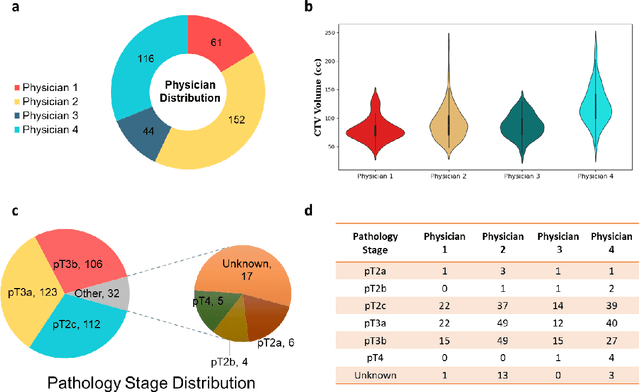
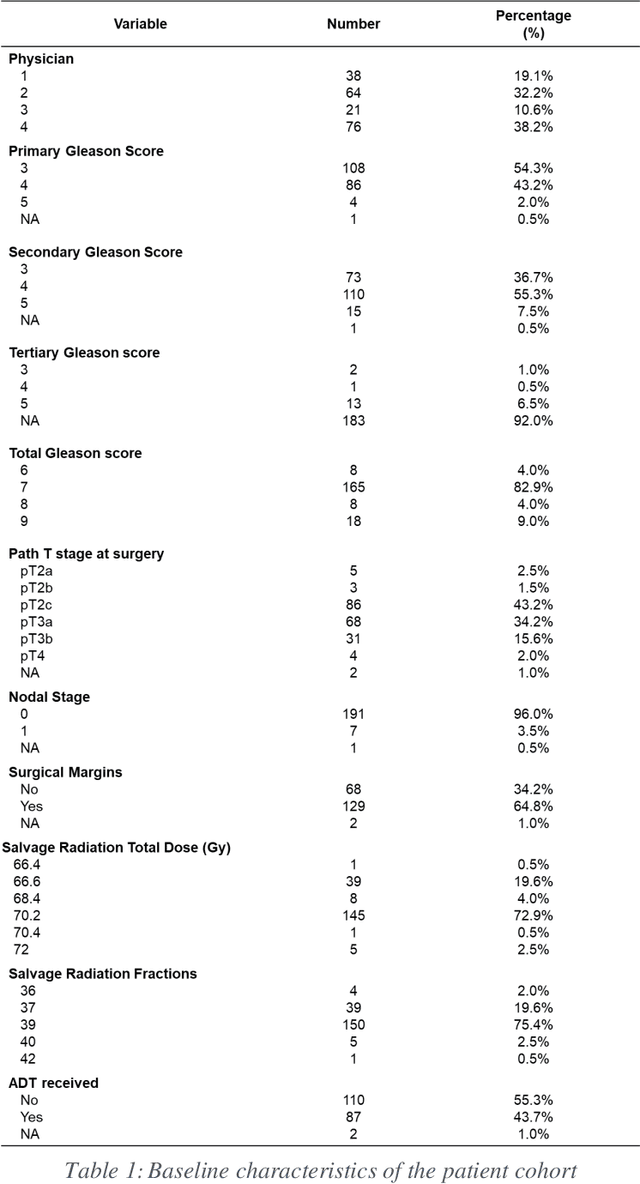
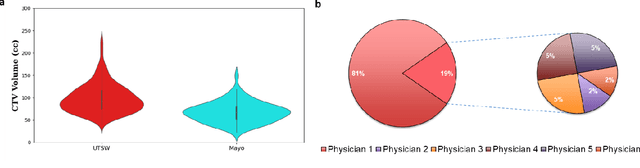
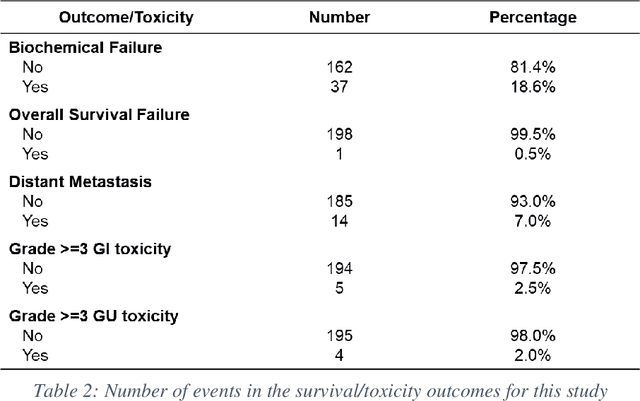
Abstract:Automatic segmentation of medical images with DL algorithms has proven to be highly successful. With most of these algorithms, inter-observer variation is an acknowledged problem, leading to sub-optimal results. This problem is even more significant in post-operative clinical target volume (post-op CTV) segmentation due to the absence of macroscopic visual tumor in the image. This study, using post-op CTV segmentation as the test bed, tries to determine if physician styles are consistent and learnable, if there is an impact of physician styles on treatment outcome and toxicity; and how to explicitly deal with physician styles in DL algorithms to facilitate its clinical acceptance. A classifier is trained to identify which physician has contoured the CTV from just the contour and corresponding CT scan, to determine if physician styles are consistent and learnable. Next, we evaluate if adapting automatic segmentation to physician styles would be clinically feasible based on a lack of difference between outcomes. For modeling different physician styles of CTV segmentation, a concept called physician style-aware (PSA) segmentation is proposed which is an encoder-multidecoder network trained with perceptual loss. With the proposed physician style-aware network (PSA-Net), Dice similarity coefficient (DSC) accuracy increases on an average of 3.4% for all physicians from a general model that is not style adapted. We show that stylistic contouring variations also exist between institutions that follow the same segmentation guidelines and show the effectiveness of the proposed method in adapting to new institutional styles. We observed an accuracy improvement of 5% in terms of DSC when adapting to the style of a separate institution.
Dosimetric impact of physician style variations in contouring CTV for post-operative prostate cancer: A deep learning based simulation study
Feb 01, 2021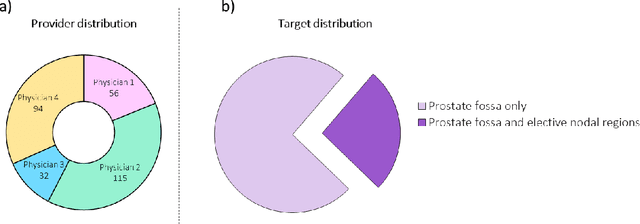
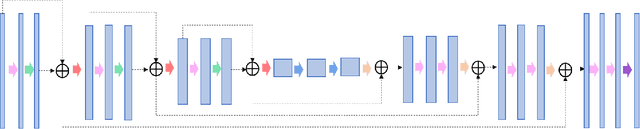
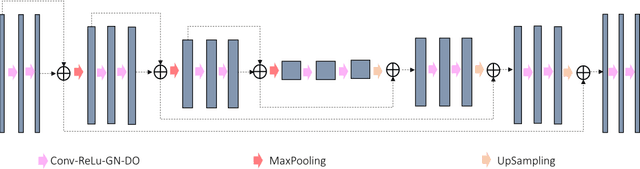
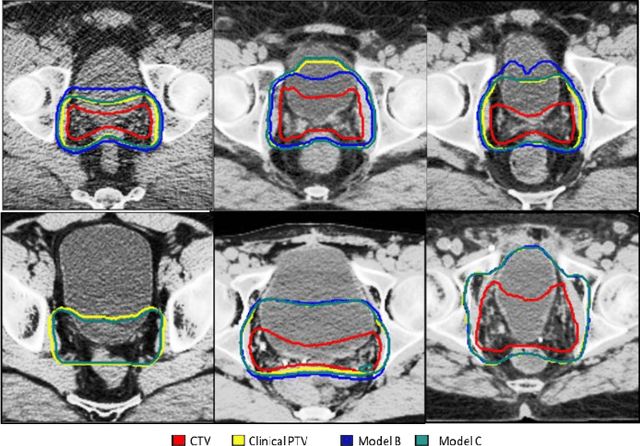
Abstract:In tumor segmentation, inter-observer variation is acknowledged to be a significant problem. This is even more significant in clinical target volume (CTV) segmentation, specifically, in post-operative settings, where a gross tumor does not exist. In this scenario, CTV is not an anatomically established structure but rather one determined by the physician based on the clinical guideline used, the preferred trade off between tumor control and toxicity, their experience, training background etc... This results in high inter-observer variability between physicians. Inter-observer variability has been considered an issue, however its dosimetric consequence is still unclear, due to the absence of multiple physician CTV contours for each patient and the significant amount of time required for dose planning. In this study, we analyze the impact that these physician stylistic variations have on organs-at-risk (OAR) dose by simulating the clinical workflow using deep learning. For a given patient previously treated by one physician, we use DL-based tools to simulate how other physicians would contour the CTV and how the corresponding dose distributions should look like for this patient. To simulate multiple physician styles, we use a previously developed in-house CTV segmentation model that can produce physician style-aware segmentations. The corresponding dose distribution is predicted using another in-house deep learning tool, which, averaging across all structures, is capable of predicting dose within 3% of the prescription dose on the test data. For every test patient, four different physician-style CTVs are considered and four different dose distributions are analyzed. OAR dose metrics are compared, showing that even though physician style variations results in organs getting different doses, all the important dose metrics except Maximum Dose point are within the clinically acceptable limit.
A deep learning-based framework for segmenting invisible clinical target volumes with estimated uncertainties for post-operative prostate cancer radiotherapy
Apr 28, 2020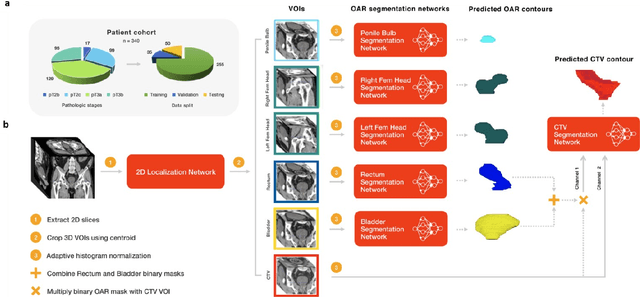
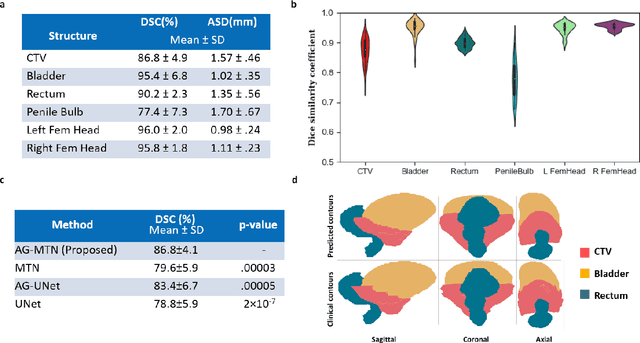
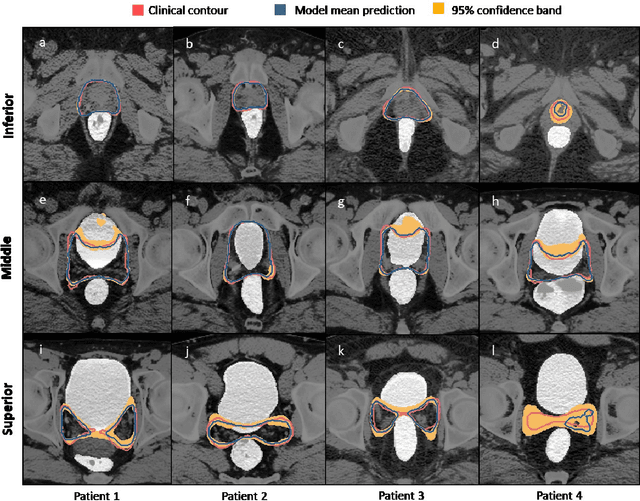
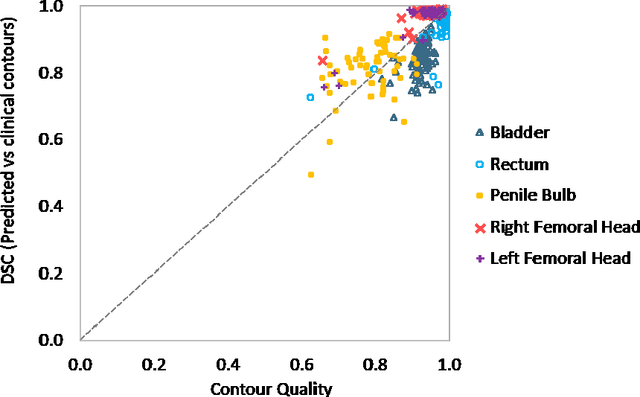
Abstract:In post-operative radiotherapy for prostate cancer, the cancerous prostate gland has been surgically removed, so the clinical target volume (CTV) to be irradiated encompasses the microscopic spread of tumor cells, which cannot be visualized in typical clinical images such as computed tomography or magnetic resonance imaging. In current clinical practice, physicians segment CTVs manually based on their relationship with nearby organs and other clinical information, per clinical guidelines. Automating post-operative prostate CTV segmentation with traditional image segmentation methods has been a major challenge. Here, we propose a deep learning model to overcome this problem by segmenting nearby organs first, then using their relationship with the CTV to assist CTV segmentation. The model proposed is trained using labels clinically approved and used for patient treatment, which are subject to relatively large inter-physician variations due to the absence of a visual ground truth. The model achieves an average Dice similarity coefficient (DSC) of 0.87 on a holdout dataset of 50 patients, much better than established methods, such as atlas-based methods (DSC<0.7). The uncertainties associated with automatically segmented CTV contours are also estimated to help physicians inspect and revise the contours, especially in areas with large inter-physician variations. We also use a 4-point grading system to show that the clinical quality of the automatically segmented CTV contours is equal to that of approved clinical contours manually drawn by physicians.
Fully Automated Organ Segmentation in Male Pelvic CT Images
May 31, 2018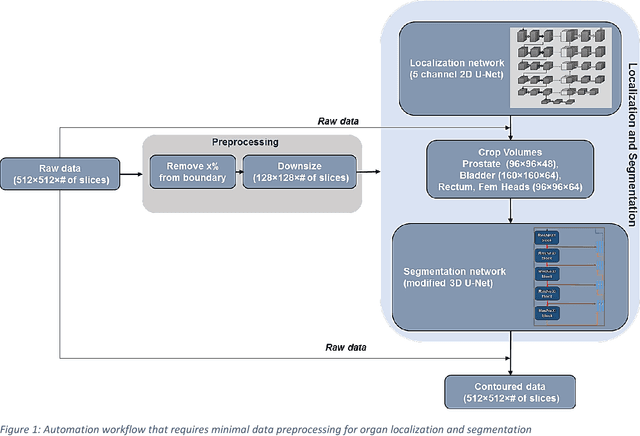
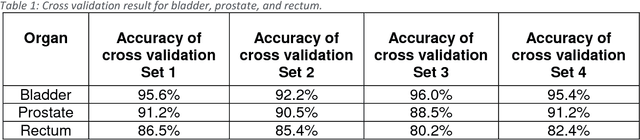
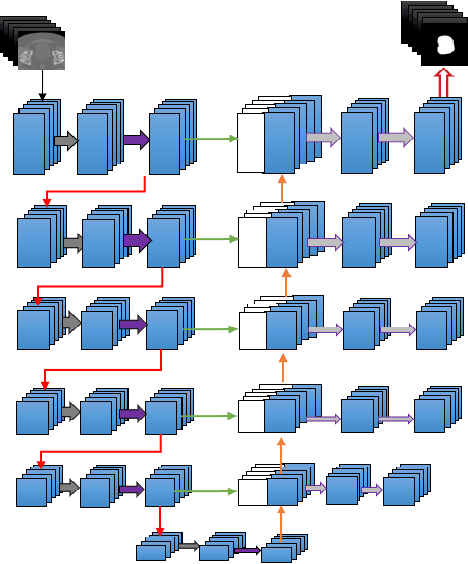
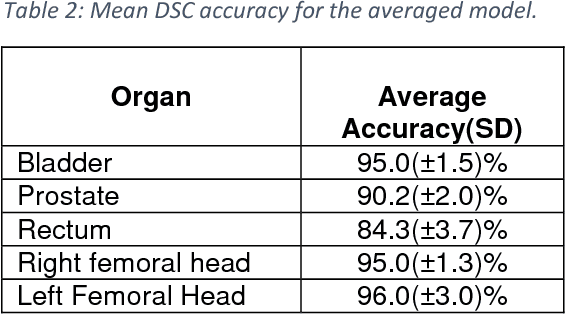
Abstract:Accurate segmentation of prostate and surrounding organs at risk is important for prostate cancer radiotherapy treatment planning. We present a fully automated workflow for male pelvic CT image segmentation using deep learning. The architecture consists of a 2D localization network followed by a 3D segmentation network for volumetric segmentation of prostate, bladder, rectum, and femoral heads. We used a multi-channel 2D U-Net followed by a 3D U-Net with encoding arm modified with aggregated residual networks, known as ResNeXt. The models were trained and tested on a pelvic CT image dataset comprising 136 patients. Test results show that 3D U-Net based segmentation achieves mean (SD) Dice coefficient values of 90 (2.0)% ,96 (3.0)%, 95 (1.3)%, 95 (1.5)%, and 84 (3.7)% for prostate, left femoral head, right femoral head, bladder, and rectum, respectively, using the proposed fully automated segmentation method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge