Jun Tan
Deep Learning Predicts Biomarker Status and Discovers Related Histomorphology Characteristics for Low-Grade Glioma
Oct 11, 2023



Abstract:Biomarker detection is an indispensable part in the diagnosis and treatment of low-grade glioma (LGG). However, current LGG biomarker detection methods rely on expensive and complex molecular genetic testing, for which professionals are required to analyze the results, and intra-rater variability is often reported. To overcome these challenges, we propose an interpretable deep learning pipeline, a Multi-Biomarker Histomorphology Discoverer (Multi-Beholder) model based on the multiple instance learning (MIL) framework, to predict the status of five biomarkers in LGG using only hematoxylin and eosin-stained whole slide images and slide-level biomarker status labels. Specifically, by incorporating the one-class classification into the MIL framework, accurate instance pseudo-labeling is realized for instance-level supervision, which greatly complements the slide-level labels and improves the biomarker prediction performance. Multi-Beholder demonstrates superior prediction performance and generalizability for five LGG biomarkers (AUROC=0.6469-0.9735) in two cohorts (n=607) with diverse races and scanning protocols. Moreover, the excellent interpretability of Multi-Beholder allows for discovering the quantitative and qualitative correlations between biomarker status and histomorphology characteristics. Our pipeline not only provides a novel approach for biomarker prediction, enhancing the applicability of molecular treatments for LGG patients but also facilitates the discovery of new mechanisms in molecular functionality and LGG progression.
A Proof-of-Concept Study of Artificial Intelligence Assisted Contour Revision
Jul 28, 2021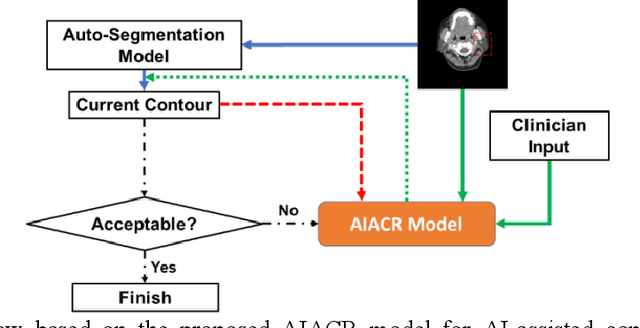

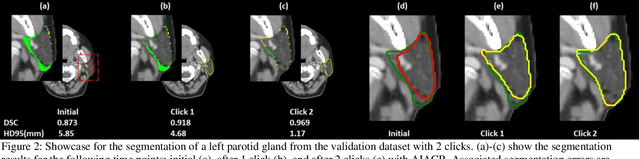
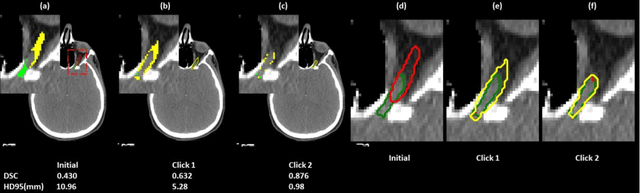
Abstract:Automatic segmentation of anatomical structures is critical for many medical applications. However, the results are not always clinically acceptable and require tedious manual revision. Here, we present a novel concept called artificial intelligence assisted contour revision (AIACR) and demonstrate its feasibility. The proposed clinical workflow of AIACR is as follows given an initial contour that requires a clinicians revision, the clinician indicates where a large revision is needed, and a trained deep learning (DL) model takes this input to update the contour. This process repeats until a clinically acceptable contour is achieved. The DL model is designed to minimize the clinicians input at each iteration and to minimize the number of iterations needed to reach acceptance. In this proof-of-concept study, we demonstrated the concept on 2D axial images of three head-and-neck cancer datasets, with the clinicians input at each iteration being one mouse click on the desired location of the contour segment. The performance of the model is quantified with Dice Similarity Coefficient (DSC) and 95th percentile of Hausdorff Distance (HD95). The average DSC/HD95 (mm) of the auto-generated initial contours were 0.82/4.3, 0.73/5.6 and 0.67/11.4 for three datasets, which were improved to 0.91/2.1, 0.86/2.4 and 0.86/4.7 with three mouse clicks, respectively. Each DL-based contour update requires around 20 ms. We proposed a novel AIACR concept that uses DL models to assist clinicians in revising contours in an efficient and effective way, and we demonstrated its feasibility by using 2D axial CT images from three head-and-neck cancer datasets.
Deep learning-based COVID-19 pneumonia classification using chest CT images: model generalizability
Feb 18, 2021



Abstract:Since the outbreak of the COVID-19 pandemic, worldwide research efforts have focused on using artificial intelligence (AI) technologies on various medical data of COVID-19-positive patients in order to identify or classify various aspects of the disease, with promising reported results. However, concerns have been raised over their generalizability, given the heterogeneous factors in training datasets. This study aims to examine the severity of this problem by evaluating deep learning (DL) classification models trained to identify COVID-19-positive patients on 3D computed tomography (CT) datasets from different countries. We collected one dataset at UT Southwestern (UTSW), and three external datasets from different countries: CC-CCII Dataset (China), COVID-CTset (Iran), and MosMedData (Russia). We divided the data into 2 classes: COVID-19-positive and COVID-19-negative patients. We trained nine identical DL-based classification models by using combinations of the datasets with a 72% train, 8% validation, and 20% test data split. The models trained on a single dataset achieved accuracy/area under the receiver operating characteristics curve (AUC) values of 0.87/0.826 (UTSW), 0.97/0.988 (CC-CCCI), and 0.86/0.873 (COVID-CTset) when evaluated on their own dataset. The models trained on multiple datasets and evaluated on a test set from one of the datasets used for training performed better. However, the performance dropped close to an AUC of 0.5 (random guess) for all models when evaluated on a different dataset outside of its training datasets. Including the MosMedData, which only contained positive labels, into the training did not necessarily help the performance on the other datasets. Multiple factors likely contribute to these results, such as patient demographics and differences in image acquisition or reconstruction, causing a data shift among different study cohorts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge