Yiyang Lin
IRS: Instance-Level 3D Scene Graphs via Room Prior Guided LiDAR-Camera Fusion
Jun 07, 2025Abstract:Indoor scene understanding remains a fundamental challenge in robotics, with direct implications for downstream tasks such as navigation and manipulation. Traditional approaches often rely on closed-set recognition or loop closure, limiting their adaptability in open-world environments. With the advent of visual foundation models (VFMs), open-vocabulary recognition and natural language querying have become feasible, unlocking new possibilities for 3D scene graph construction. In this paper, we propose a robust and efficient framework for instance-level 3D scene graph construction via LiDAR-camera fusion. Leveraging LiDAR's wide field of view (FOV) and long-range sensing capabilities, we rapidly acquire room-level geometric priors. Multi-level VFMs are employed to improve the accuracy and consistency of semantic extraction. During instance fusion, room-based segmentation enables parallel processing, while the integration of geometric and semantic cues significantly enhances fusion accuracy and robustness. Compared to state-of-the-art methods, our approach achieves up to an order-of-magnitude improvement in construction speed while maintaining high semantic precision. Extensive experiments in both simulated and real-world environments validate the effectiveness of our approach. We further demonstrate its practical value through a language-guided semantic navigation task, highlighting its potential for real-world robotic applications.
ConcealGS: Concealing Invisible Copyright Information in 3D Gaussian Splatting
Jan 07, 2025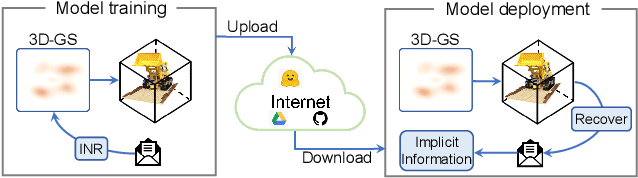
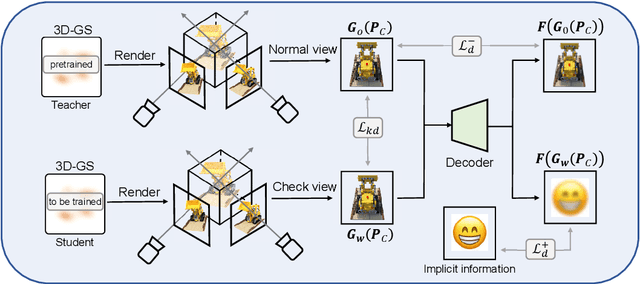
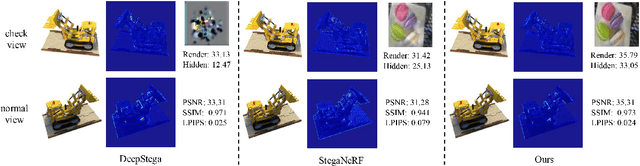
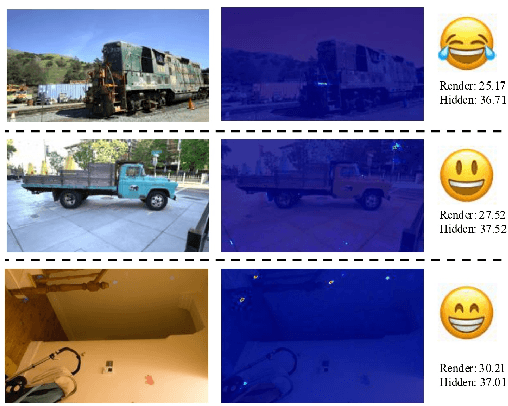
Abstract:With the rapid development of 3D reconstruction technology, the widespread distribution of 3D data has become a future trend. While traditional visual data (such as images and videos) and NeRF-based formats already have mature techniques for copyright protection, steganographic techniques for the emerging 3D Gaussian Splatting (3D-GS) format have yet to be fully explored. To address this, we propose ConcealGS, an innovative method for embedding implicit information into 3D-GS. By introducing the knowledge distillation and gradient optimization strategy based on 3D-GS, ConcealGS overcomes the limitations of NeRF-based models and enhances the robustness of implicit information and the quality of 3D reconstruction. We evaluate ConcealGS in various potential application scenarios, and experimental results have demonstrated that ConcealGS not only successfully recovers implicit information but also has almost no impact on rendering quality, providing a new approach for embedding invisible and recoverable information into 3D models in the future.
AV-GAN: Attention-Based Varifocal Generative Adversarial Network for Uneven Medical Image Translation
Apr 16, 2024Abstract:Different types of staining highlight different structures in organs, thereby assisting in diagnosis. However, due to the impossibility of repeated staining, we cannot obtain different types of stained slides of the same tissue area. Translating the slide that is easy to obtain (e.g., H&E) to slides of staining types difficult to obtain (e.g., MT, PAS) is a promising way to solve this problem. However, some regions are closely connected to other regions, and to maintain this connection, they often have complex structures and are difficult to translate, which may lead to wrong translations. In this paper, we propose the Attention-Based Varifocal Generative Adversarial Network (AV-GAN), which solves multiple problems in pathologic image translation tasks, such as uneven translation difficulty in different regions, mutual interference of multiple resolution information, and nuclear deformation. Specifically, we develop an Attention-Based Key Region Selection Module, which can attend to regions with higher translation difficulty. We then develop a Varifocal Module to translate these regions at multiple resolutions. Experimental results show that our proposed AV-GAN outperforms existing image translation methods with two virtual kidney tissue staining tasks and improves FID values by 15.9 and 4.16 respectively in the H&E-MT and H&E-PAS tasks.
Deep Learning Predicts Biomarker Status and Discovers Related Histomorphology Characteristics for Low-Grade Glioma
Oct 11, 2023



Abstract:Biomarker detection is an indispensable part in the diagnosis and treatment of low-grade glioma (LGG). However, current LGG biomarker detection methods rely on expensive and complex molecular genetic testing, for which professionals are required to analyze the results, and intra-rater variability is often reported. To overcome these challenges, we propose an interpretable deep learning pipeline, a Multi-Biomarker Histomorphology Discoverer (Multi-Beholder) model based on the multiple instance learning (MIL) framework, to predict the status of five biomarkers in LGG using only hematoxylin and eosin-stained whole slide images and slide-level biomarker status labels. Specifically, by incorporating the one-class classification into the MIL framework, accurate instance pseudo-labeling is realized for instance-level supervision, which greatly complements the slide-level labels and improves the biomarker prediction performance. Multi-Beholder demonstrates superior prediction performance and generalizability for five LGG biomarkers (AUROC=0.6469-0.9735) in two cohorts (n=607) with diverse races and scanning protocols. Moreover, the excellent interpretability of Multi-Beholder allows for discovering the quantitative and qualitative correlations between biomarker status and histomorphology characteristics. Our pipeline not only provides a novel approach for biomarker prediction, enhancing the applicability of molecular treatments for LGG patients but also facilitates the discovery of new mechanisms in molecular functionality and LGG progression.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge