Yixuan Yuan
MedSAM-Agent: Empowering Interactive Medical Image Segmentation with Multi-turn Agentic Reinforcement Learning
Feb 03, 2026Abstract:Medical image segmentation is evolving from task-specific models toward generalizable frameworks. Recent research leverages Multi-modal Large Language Models (MLLMs) as autonomous agents, employing reinforcement learning with verifiable reward (RLVR) to orchestrate specialized tools like the Segment Anything Model (SAM). However, these approaches often rely on single-turn, rigid interaction strategies and lack process-level supervision during training, which hinders their ability to fully exploit the dynamic potential of interactive tools and leads to redundant actions. To bridge this gap, we propose MedSAM-Agent, a framework that reformulates interactive segmentation as a multi-step autonomous decision-making process. First, we introduce a hybrid prompting strategy for expert-curated trajectory generation, enabling the model to internalize human-like decision heuristics and adaptive refinement strategies. Furthermore, we develop a two-stage training pipeline that integrates multi-turn, end-to-end outcome verification with a clinical-fidelity process reward design to promote interaction parsimony and decision efficiency. Extensive experiments across 6 medical modalities and 21 datasets demonstrate that MedSAM-Agent achieves state-of-the-art performance, effectively unifying autonomous medical reasoning with robust, iterative optimization. Code is available \href{https://github.com/CUHK-AIM-Group/MedSAM-Agent}{here}.
VDE Bench: Evaluating The Capability of Image Editing Models to Modify Visual Documents
Jan 27, 2026Abstract:In recent years, multimodal image editing models have achieved substantial progress, enabling users to manipulate visual content through natural language in a flexible and interactive manner. Nevertheless, an important yet insufficiently explored research direction remains visual document image editing, which involves modifying textual content within images while faithfully preserving the original text style and background context. Existing approaches, including AnyText, GlyphControl, and TextCtrl, predominantly focus on English-language scenarios and documents with relatively sparse textual layouts, thereby failing to adequately address dense, structurally complex documents or non-Latin scripts such as Chinese. To bridge this gap, we propose \textbf{V}isual \textbf{D}oc \textbf{E}dit Bench(VDE Bench), a rigorously human-annotated and evaluated benchmark specifically designed to assess image editing models on multilingual and complex visual document editing tasks. The benchmark comprises a high-quality dataset encompassing densely textual documents in both English and Chinese, including academic papers, posters, presentation slides, examination materials, and newspapers. Furthermore, we introduce a decoupled evaluation framework that systematically quantifies editing performance at the OCR parsing level, enabling fine-grained assessment of text modification accuracy. Based on this benchmark, we conduct a comprehensive evaluation of representative state-of-the-art image editing models. Manual verification demonstrates a strong consistency between human judgments and automated evaluation metrics. VDE Bench constitutes the first systematic benchmark for evaluating image editing models on multilingual and densely textual visual documents.
Unveiling and Bridging the Functional Perception Gap in MLLMs: Atomic Visual Alignment and Hierarchical Evaluation via PET-Bench
Jan 06, 2026Abstract:While Multimodal Large Language Models (MLLMs) have demonstrated remarkable proficiency in tasks such as abnormality detection and report generation for anatomical modalities, their capability in functional imaging remains largely unexplored. In this work, we identify and quantify a fundamental functional perception gap: the inability of current vision encoders to decode functional tracer biodistribution independent of morphological priors. Identifying Positron Emission Tomography (PET) as the quintessential modality to investigate this disconnect, we introduce PET-Bench, the first large-scale functional imaging benchmark comprising 52,308 hierarchical QA pairs from 9,732 multi-site, multi-tracer PET studies. Extensive evaluation of 19 state-of-the-art MLLMs reveals a critical safety hazard termed the Chain-of-Thought (CoT) hallucination trap. We observe that standard CoT prompting, widely considered to enhance reasoning, paradoxically decouples linguistic generation from visual evidence in PET, producing clinically fluent but factually ungrounded diagnoses. To resolve this, we propose Atomic Visual Alignment (AVA), a simple fine-tuning strategy that enforces the mastery of low-level functional perception prior to high-level diagnostic reasoning. Our results demonstrate that AVA effectively bridges the perception gap, transforming CoT from a source of hallucination into a robust inference tool and improving diagnostic accuracy by up to 14.83%. Code and data are available at https://github.com/yezanting/PET-Bench.
Medical Reasoning in the Era of LLMs: A Systematic Review of Enhancement Techniques and Applications
Aug 01, 2025Abstract:The proliferation of Large Language Models (LLMs) in medicine has enabled impressive capabilities, yet a critical gap remains in their ability to perform systematic, transparent, and verifiable reasoning, a cornerstone of clinical practice. This has catalyzed a shift from single-step answer generation to the development of LLMs explicitly designed for medical reasoning. This paper provides the first systematic review of this emerging field. We propose a taxonomy of reasoning enhancement techniques, categorized into training-time strategies (e.g., supervised fine-tuning, reinforcement learning) and test-time mechanisms (e.g., prompt engineering, multi-agent systems). We analyze how these techniques are applied across different data modalities (text, image, code) and in key clinical applications such as diagnosis, education, and treatment planning. Furthermore, we survey the evolution of evaluation benchmarks from simple accuracy metrics to sophisticated assessments of reasoning quality and visual interpretability. Based on an analysis of 60 seminal studies from 2022-2025, we conclude by identifying critical challenges, including the faithfulness-plausibility gap and the need for native multimodal reasoning, and outlining future directions toward building efficient, robust, and sociotechnically responsible medical AI.
EndoGen: Conditional Autoregressive Endoscopic Video Generation
Jul 23, 2025Abstract:Endoscopic video generation is crucial for advancing medical imaging and enhancing diagnostic capabilities. However, prior efforts in this field have either focused on static images, lacking the dynamic context required for practical applications, or have relied on unconditional generation that fails to provide meaningful references for clinicians. Therefore, in this paper, we propose the first conditional endoscopic video generation framework, namely EndoGen. Specifically, we build an autoregressive model with a tailored Spatiotemporal Grid-Frame Patterning (SGP) strategy. It reformulates the learning of generating multiple frames as a grid-based image generation pattern, which effectively capitalizes the inherent global dependency modeling capabilities of autoregressive architectures. Furthermore, we propose a Semantic-Aware Token Masking (SAT) mechanism, which enhances the model's ability to produce rich and diverse content by selectively focusing on semantically meaningful regions during the generation process. Through extensive experiments, we demonstrate the effectiveness of our framework in generating high-quality, conditionally guided endoscopic content, and improves the performance of downstream task of polyp segmentation. Code released at https://www.github.com/CUHK-AIM-Group/EndoGen.
Age Sensitive Hippocampal Functional Connectivity: New Insights from 3D CNNs and Saliency Mapping
Jul 02, 2025Abstract:Grey matter loss in the hippocampus is a hallmark of neurobiological aging, yet understanding the corresponding changes in its functional connectivity remains limited. Seed-based functional connectivity (FC) analysis enables voxel-wise mapping of the hippocampus's synchronous activity with cortical regions, offering a window into functional reorganization during aging. In this study, we develop an interpretable deep learning framework to predict brain age from hippocampal FC using a three-dimensional convolutional neural network (3D CNN) combined with LayerCAM saliency mapping. This approach maps key hippocampal-cortical connections, particularly with the precuneus, cuneus, posterior cingulate cortex, parahippocampal cortex, left superior parietal lobule, and right superior temporal sulcus, that are highly sensitive to age. Critically, disaggregating anterior and posterior hippocampal FC reveals distinct mapping aligned with their known functional specializations. These findings provide new insights into the functional mechanisms of hippocampal aging and demonstrate the power of explainable deep learning to uncover biologically meaningful patterns in neuroimaging data.
RadFabric: Agentic AI System with Reasoning Capability for Radiology
Jun 17, 2025Abstract:Chest X ray (CXR) imaging remains a critical diagnostic tool for thoracic conditions, but current automated systems face limitations in pathology coverage, diagnostic accuracy, and integration of visual and textual reasoning. To address these gaps, we propose RadFabric, a multi agent, multimodal reasoning framework that unifies visual and textual analysis for comprehensive CXR interpretation. RadFabric is built on the Model Context Protocol (MCP), enabling modularity, interoperability, and scalability for seamless integration of new diagnostic agents. The system employs specialized CXR agents for pathology detection, an Anatomical Interpretation Agent to map visual findings to precise anatomical structures, and a Reasoning Agent powered by large multimodal reasoning models to synthesize visual, anatomical, and clinical data into transparent and evidence based diagnoses. RadFabric achieves significant performance improvements, with near-perfect detection of challenging pathologies like fractures (1.000 accuracy) and superior overall diagnostic accuracy (0.799) compared to traditional systems (0.229 to 0.527). By integrating cross modal feature alignment and preference-driven reasoning, RadFabric advances AI-driven radiology toward transparent, anatomically precise, and clinically actionable CXR analysis.
Voxel-Level Brain States Prediction Using Swin Transformer
Jun 13, 2025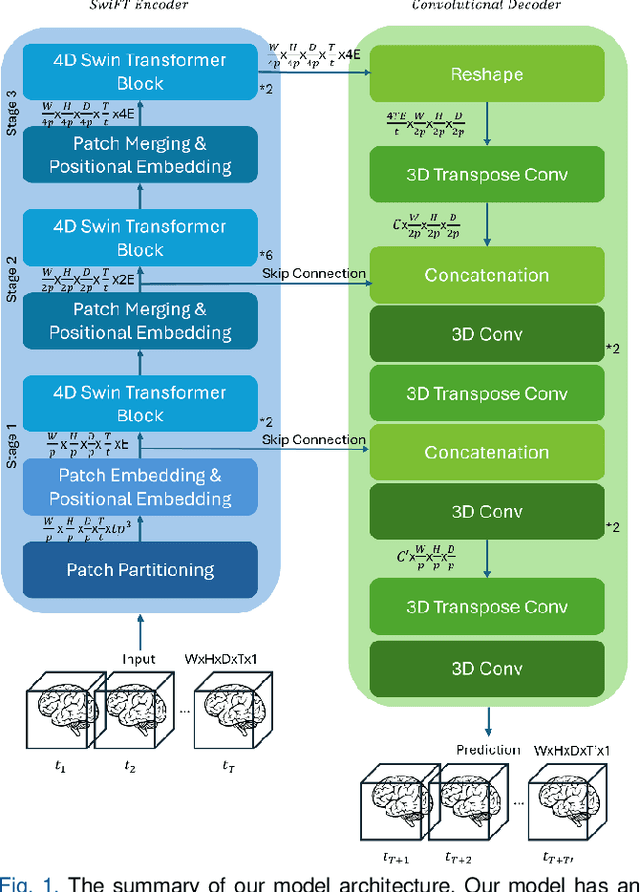
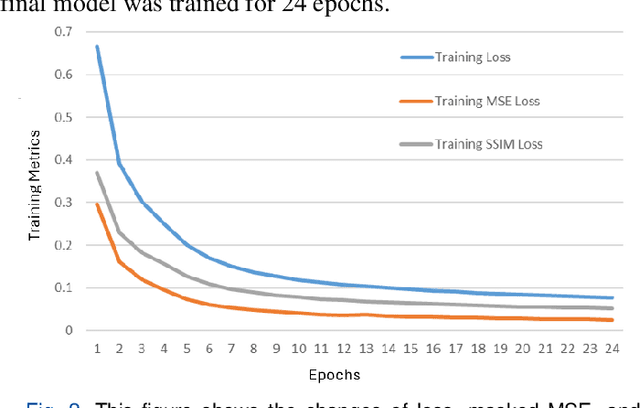
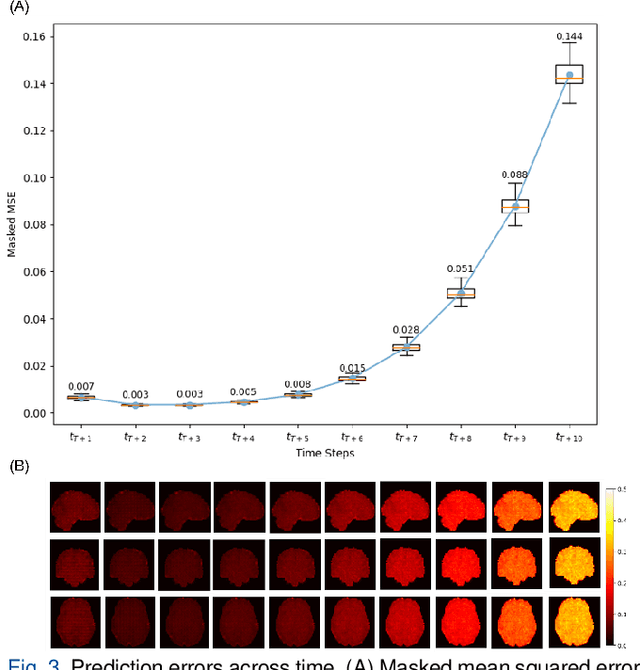
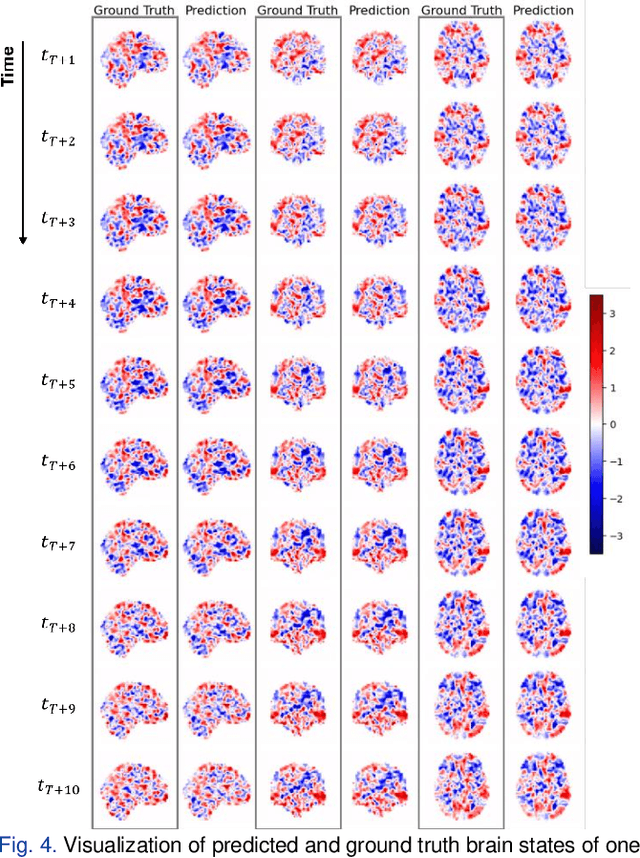
Abstract:Understanding brain dynamics is important for neuroscience and mental health. Functional magnetic resonance imaging (fMRI) enables the measurement of neural activities through blood-oxygen-level-dependent (BOLD) signals, which represent brain states. In this study, we aim to predict future human resting brain states with fMRI. Due to the 3D voxel-wise spatial organization and temporal dependencies of the fMRI data, we propose a novel architecture which employs a 4D Shifted Window (Swin) Transformer as encoder to efficiently learn spatio-temporal information and a convolutional decoder to enable brain state prediction at the same spatial and temporal resolution as the input fMRI data. We used 100 unrelated subjects from the Human Connectome Project (HCP) for model training and testing. Our novel model has shown high accuracy when predicting 7.2s resting-state brain activities based on the prior 23.04s fMRI time series. The predicted brain states highly resemble BOLD contrast and dynamics. This work shows promising evidence that the spatiotemporal organization of the human brain can be learned by a Swin Transformer model, at high resolution, which provides a potential for reducing the fMRI scan time and the development of brain-computer interfaces in the future.
Towards a general-purpose foundation model for fMRI analysis
Jun 11, 2025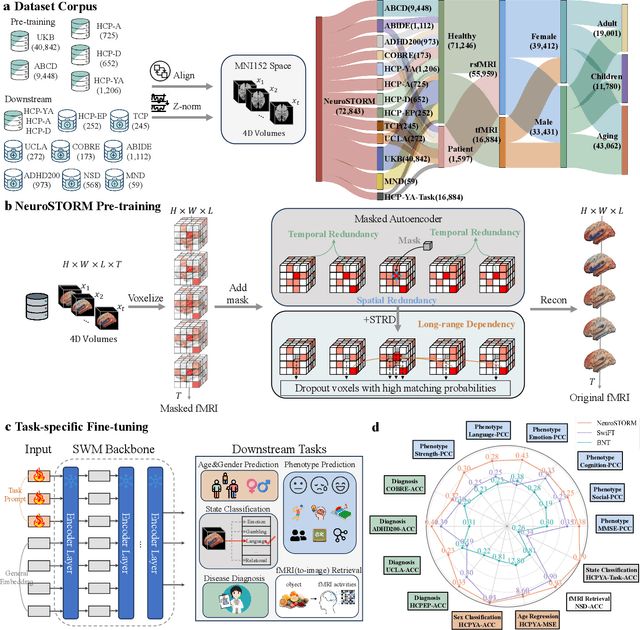

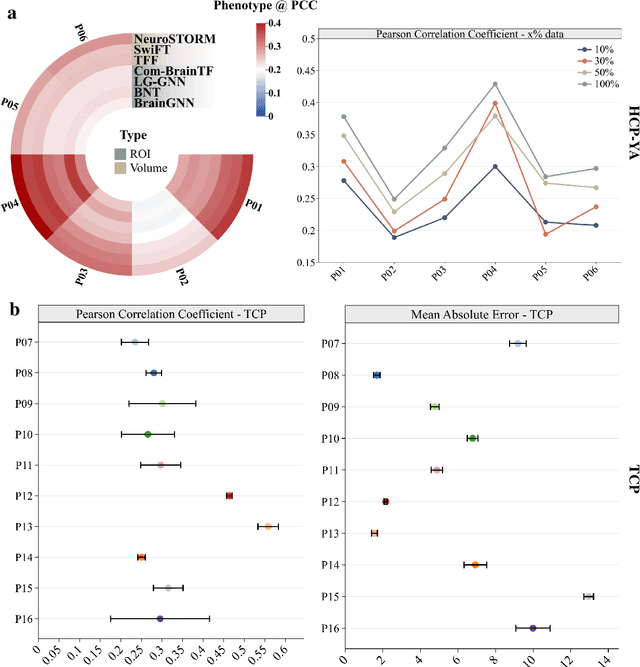
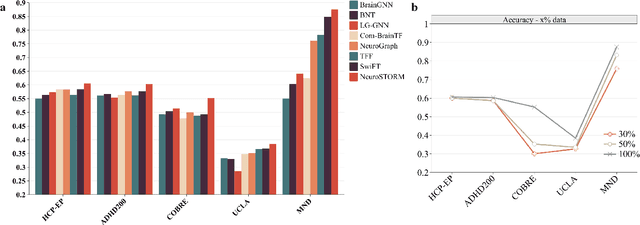
Abstract:Functional Magnetic Resonance Imaging (fMRI) is essential for studying brain function and diagnosing neurological disorders, but current analysis methods face reproducibility and transferability issues due to complex pre-processing and task-specific models. We introduce NeuroSTORM (Neuroimaging Foundation Model with Spatial-Temporal Optimized Representation Modeling), a generalizable framework that directly learns from 4D fMRI volumes and enables efficient knowledge transfer across diverse applications. NeuroSTORM is pre-trained on 28.65 million fMRI frames (>9,000 hours) from over 50,000 subjects across multiple centers and ages 5 to 100. Using a Mamba backbone and a shifted scanning strategy, it efficiently processes full 4D volumes. We also propose a spatial-temporal optimized pre-training approach and task-specific prompt tuning to improve transferability. NeuroSTORM outperforms existing methods across five tasks: age/gender prediction, phenotype prediction, disease diagnosis, fMRI-to-image retrieval, and task-based fMRI classification. It demonstrates strong clinical utility on datasets from hospitals in the U.S., South Korea, and Australia, achieving top performance in disease diagnosis and cognitive phenotype prediction. NeuroSTORM provides a standardized, open-source foundation model to improve reproducibility and transferability in fMRI-based clinical research.
Track Any Anomalous Object: A Granular Video Anomaly Detection Pipeline
Jun 05, 2025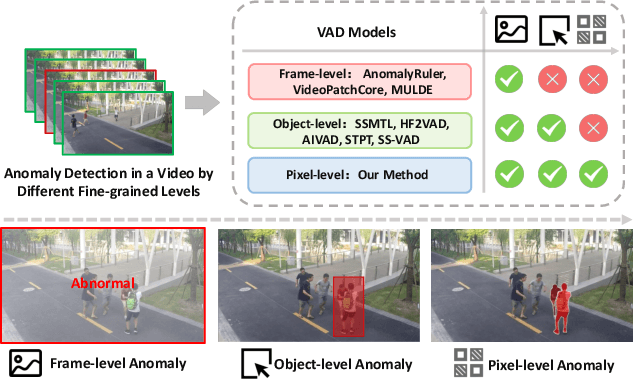
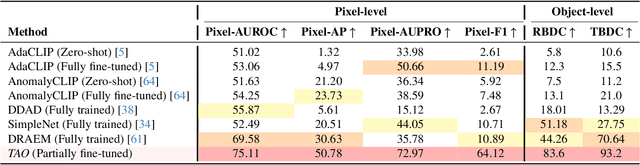
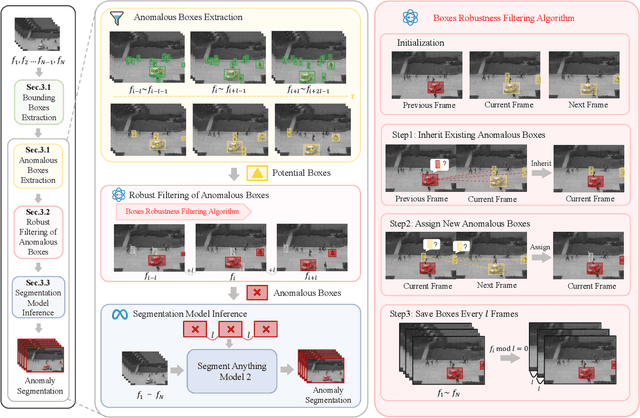

Abstract:Video anomaly detection (VAD) is crucial in scenarios such as surveillance and autonomous driving, where timely detection of unexpected activities is essential. Although existing methods have primarily focused on detecting anomalous objects in videos -- either by identifying anomalous frames or objects -- they often neglect finer-grained analysis, such as anomalous pixels, which limits their ability to capture a broader range of anomalies. To address this challenge, we propose a new framework called Track Any Anomalous Object (TAO), which introduces a granular video anomaly detection pipeline that, for the first time, integrates the detection of multiple fine-grained anomalous objects into a unified framework. Unlike methods that assign anomaly scores to every pixel, our approach transforms the problem into pixel-level tracking of anomalous objects. By linking anomaly scores to downstream tasks such as segmentation and tracking, our method removes the need for threshold tuning and achieves more precise anomaly localization in long and complex video sequences. Experiments demonstrate that TAO sets new benchmarks in accuracy and robustness. Project page available online.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge