Qiushi Yang
MoSAM: Motion-Guided Segment Anything Model with Spatial-Temporal Memory Selection
Apr 30, 2025



Abstract:The recent Segment Anything Model 2 (SAM2) has demonstrated exceptional capabilities in interactive object segmentation for both images and videos. However, as a foundational model on interactive segmentation, SAM2 performs segmentation directly based on mask memory from the past six frames, leading to two significant challenges. Firstly, during inference in videos, objects may disappear since SAM2 relies solely on memory without accounting for object motion information, which limits its long-range object tracking capabilities. Secondly, its memory is constructed from fixed past frames, making it susceptible to challenges associated with object disappearance or occlusion, due to potentially inaccurate segmentation results in memory. To address these problems, we present MoSAM, incorporating two key strategies to integrate object motion cues into the model and establish more reliable feature memory. Firstly, we propose Motion-Guided Prompting (MGP), which represents the object motion in both sparse and dense manners, then injects them into SAM2 through a set of motion-guided prompts. MGP enables the model to adjust its focus towards the direction of motion, thereby enhancing the object tracking capabilities. Furthermore, acknowledging that past segmentation results may be inaccurate, we devise a Spatial-Temporal Memory Selection (ST-MS) mechanism that dynamically identifies frames likely to contain accurate segmentation in both pixel- and frame-level. By eliminating potentially inaccurate mask predictions from memory, we can leverage more reliable memory features to exploit similar regions for improving segmentation results. Extensive experiments on various benchmarks of video object segmentation and video instance segmentation demonstrate that our MoSAM achieves state-of-the-art results compared to other competitors.
FedBM: Stealing Knowledge from Pre-trained Language Models for Heterogeneous Federated Learning
Feb 24, 2025Abstract:Federated learning (FL) has shown great potential in medical image computing since it provides a decentralized learning paradigm that allows multiple clients to train a model collaboratively without privacy leakage. However, current studies have shown that data heterogeneity incurs local learning bias in classifiers and feature extractors of client models during local training, leading to the performance degradation of a federation system. To address these issues, we propose a novel framework called Federated Bias eliMinating (FedBM) to get rid of local learning bias in heterogeneous federated learning (FL), which mainly consists of two modules, i.e., Linguistic Knowledge-based Classifier Construction (LKCC) and Concept-guided Global Distribution Estimation (CGDE). Specifically, LKCC exploits class concepts, prompts and pre-trained language models (PLMs) to obtain concept embeddings. These embeddings are used to estimate the latent concept distribution of each class in the linguistic space. Based on the theoretical derivation, we can rely on these distributions to pre-construct a high-quality classifier for clients to achieve classification optimization, which is frozen to avoid classifier bias during local training. CGDE samples probabilistic concept embeddings from the latent concept distributions to learn a conditional generator to capture the input space of the global model. Three regularization terms are introduced to improve the quality and utility of the generator. The generator is shared by all clients and produces pseudo data to calibrate updates of local feature extractors. Extensive comparison experiments and ablation studies on public datasets demonstrate the superior performance of FedBM over state-of-the-arts and confirm the effectiveness of each module, respectively. The code is available at https://github.com/CUHK-AIM-Group/FedBM.
Long-tailed Medical Diagnosis with Relation-aware Representation Learning and Iterative Classifier Calibration
Feb 05, 2025



Abstract:Recently computer-aided diagnosis has demonstrated promising performance, effectively alleviating the workload of clinicians. However, the inherent sample imbalance among different diseases leads algorithms biased to the majority categories, leading to poor performance for rare categories. Existing works formulated this challenge as a long-tailed problem and attempted to tackle it by decoupling the feature representation and classification. Yet, due to the imbalanced distribution and limited samples from tail classes, these works are prone to biased representation learning and insufficient classifier calibration. To tackle these problems, we propose a new Long-tailed Medical Diagnosis (LMD) framework for balanced medical image classification on long-tailed datasets. In the initial stage, we develop a Relation-aware Representation Learning (RRL) scheme to boost the representation ability by encouraging the encoder to capture intrinsic semantic features through different data augmentations. In the subsequent stage, we propose an Iterative Classifier Calibration (ICC) scheme to calibrate the classifier iteratively. This is achieved by generating a large number of balanced virtual features and fine-tuning the encoder using an Expectation-Maximization manner. The proposed ICC compensates for minority categories to facilitate unbiased classifier optimization while maintaining the diagnostic knowledge in majority classes. Comprehensive experiments on three public long-tailed medical datasets demonstrate that our LMD framework significantly surpasses state-of-the-art approaches. The source code can be accessed at https://github.com/peterlipan/LMD.
Polyp-Gen: Realistic and Diverse Polyp Image Generation for Endoscopic Dataset Expansion
Jan 29, 2025



Abstract:Automated diagnostic systems (ADS) have shown significant potential in the early detection of polyps during endoscopic examinations, thereby reducing the incidence of colorectal cancer. However, due to high annotation costs and strict privacy concerns, acquiring high-quality endoscopic images poses a considerable challenge in the development of ADS. Despite recent advancements in generating synthetic images for dataset expansion, existing endoscopic image generation algorithms failed to accurately generate the details of polyp boundary regions and typically required medical priors to specify plausible locations and shapes of polyps, which limited the realism and diversity of the generated images. To address these limitations, we present Polyp-Gen, the first full-automatic diffusion-based endoscopic image generation framework. Specifically, we devise a spatial-aware diffusion training scheme with a lesion-guided loss to enhance the structural context of polyp boundary regions. Moreover, to capture medical priors for the localization of potential polyp areas, we introduce a hierarchical retrieval-based sampling strategy to match similar fine-grained spatial features. In this way, our Polyp-Gen can generate realistic and diverse endoscopic images for building reliable ADS. Extensive experiments demonstrate the state-of-the-art generation quality, and the synthetic images can improve the downstream polyp detection task. Additionally, our Polyp-Gen has shown remarkable zero-shot generalizability on other datasets. The source code is available at https://github.com/CUHK-AIM-Group/Polyp-Gen.
Focus on Focus: Focus-oriented Representation Learning and Multi-view Cross-modal Alignment for Glioma Grading
Aug 16, 2024



Abstract:Recently, multimodal deep learning, which integrates histopathology slides and molecular biomarkers, has achieved a promising performance in glioma grading. Despite great progress, due to the intra-modality complexity and inter-modality heterogeneity, existing studies suffer from inadequate histopathology representation learning and inefficient molecular-pathology knowledge alignment. These two issues hinder existing methods to precisely interpret diagnostic molecular-pathology features, thereby limiting their grading performance. Moreover, the real-world applicability of existing multimodal approaches is significantly restricted as molecular biomarkers are not always available during clinical deployment. To address these problems, we introduce a novel Focus on Focus (FoF) framework with paired pathology-genomic training and applicable pathology-only inference, enhancing molecular-pathology representation effectively. Specifically, we propose a Focus-oriented Representation Learning (FRL) module to encourage the model to identify regions positively or negatively related to glioma grading and guide it to focus on the diagnostic areas with a consistency constraint. To effectively link the molecular biomarkers to morphological features, we propose a Multi-view Cross-modal Alignment (MCA) module that projects histopathology representations into molecular subspaces, aligning morphological features with corresponding molecular biomarker status by supervised contrastive learning. Experiments on the TCGA GBM-LGG dataset demonstrate that our FoF framework significantly improves the glioma grading. Remarkably, our FoF achieves superior performance using only histopathology slides compared to existing multimodal methods. The source code is available at https://github.com/peterlipan/FoF.
Unified Multi-modal Diagnostic Framework with Reconstruction Pre-training and Heterogeneity-combat Tuning
Apr 09, 2024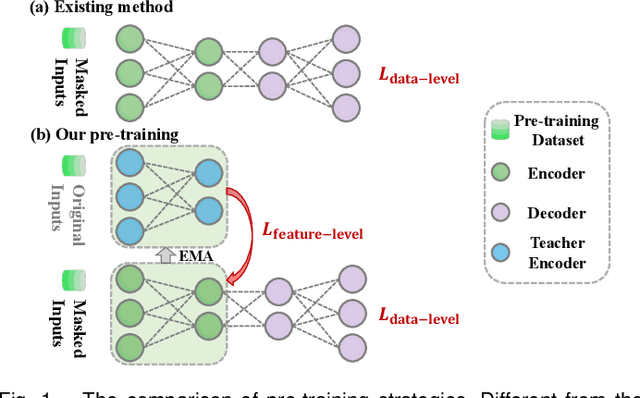
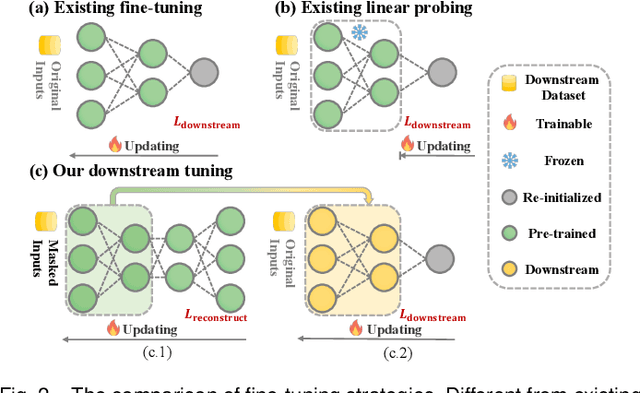
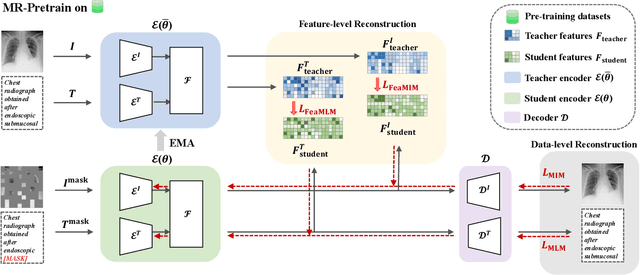
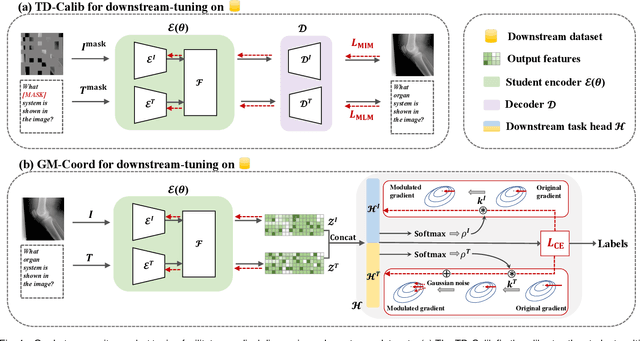
Abstract:Medical multi-modal pre-training has revealed promise in computer-aided diagnosis by leveraging large-scale unlabeled datasets. However, existing methods based on masked autoencoders mainly rely on data-level reconstruction tasks, but lack high-level semantic information. Furthermore, two significant heterogeneity challenges hinder the transfer of pre-trained knowledge to downstream tasks, \textit{i.e.}, the distribution heterogeneity between pre-training data and downstream data, and the modality heterogeneity within downstream data. To address these challenges, we propose a Unified Medical Multi-modal Diagnostic (UMD) framework with tailored pre-training and downstream tuning strategies. Specifically, to enhance the representation abilities of vision and language encoders, we propose the Multi-level Reconstruction Pre-training (MR-Pretrain) strategy, including a feature-level and data-level reconstruction, which guides models to capture the semantic information from masked inputs of different modalities. Moreover, to tackle two kinds of heterogeneities during the downstream tuning, we present the heterogeneity-combat downstream tuning strategy, which consists of a Task-oriented Distribution Calibration (TD-Calib) and a Gradient-guided Modality Coordination (GM-Coord). In particular, TD-Calib fine-tunes the pre-trained model regarding the distribution of downstream datasets, and GM-Coord adjusts the gradient weights according to the dynamic optimization status of different modalities. Extensive experiments on five public medical datasets demonstrate the effectiveness of our UMD framework, which remarkably outperforms existing approaches on three kinds of downstream tasks.
Review of Large Vision Models and Visual Prompt Engineering
Jul 03, 2023
Abstract:Visual prompt engineering is a fundamental technology in the field of visual and image Artificial General Intelligence, serving as a key component for achieving zero-shot capabilities. As the development of large vision models progresses, the importance of prompt engineering becomes increasingly evident. Designing suitable prompts for specific visual tasks has emerged as a meaningful research direction. This review aims to summarize the methods employed in the computer vision domain for large vision models and visual prompt engineering, exploring the latest advancements in visual prompt engineering. We present influential large models in the visual domain and a range of prompt engineering methods employed on these models. It is our hope that this review provides a comprehensive and systematic description of prompt engineering methods based on large visual models, offering valuable insights for future researchers in their exploration of this field.
Prompt Engineering for Healthcare: Methodologies and Applications
Apr 28, 2023Abstract:This review will introduce the latest advances in prompt engineering in the field of natural language processing (NLP) for the medical domain. First, we will provide a brief overview of the development of prompt engineering and emphasize its significant contributions to healthcare NLP applications such as question-answering systems, text summarization, and machine translation. With the continuous improvement of general large language models, the importance of prompt engineering in the healthcare domain is becoming increasingly prominent. The aim of this article is to provide useful resources and bridges for healthcare NLP researchers to better explore the application of prompt engineering in this field. We hope that this review can provide new ideas and inspire ample possibilities for research and application in medical NLP.
Towards Robust Adaptive Object Detection under Noisy Annotations
Apr 06, 2022



Abstract:Domain Adaptive Object Detection (DAOD) models a joint distribution of images and labels from an annotated source domain and learns a domain-invariant transformation to estimate the target labels with the given target domain images. Existing methods assume that the source domain labels are completely clean, yet large-scale datasets often contain error-prone annotations due to instance ambiguity, which may lead to a biased source distribution and severely degrade the performance of the domain adaptive detector de facto. In this paper, we represent the first effort to formulate noisy DAOD and propose a Noise Latent Transferability Exploration (NLTE) framework to address this issue. It is featured with 1) Potential Instance Mining (PIM), which leverages eligible proposals to recapture the miss-annotated instances from the background; 2) Morphable Graph Relation Module (MGRM), which models the adaptation feasibility and transition probability of noisy samples with relation matrices; 3) Entropy-Aware Gradient Reconcilement (EAGR), which incorporates the semantic information into the discrimination process and enforces the gradients provided by noisy and clean samples to be consistent towards learning domain-invariant representations. A thorough evaluation on benchmark DAOD datasets with noisy source annotations validates the effectiveness of NLTE. In particular, NLTE improves the mAP by 8.4\% under 60\% corrupted annotations and even approaches the ideal upper bound of training on a clean source dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge