Zhen Chen
School of Cyber Science and Technology, University of Science and Technology of China
ReWeaver: Towards Simulation-Ready and Topology-Accurate Garment Reconstruction
Jan 23, 2026Abstract:High-quality 3D garment reconstruction plays a crucial role in mitigating the sim-to-real gap in applications such as digital avatars, virtual try-on and robotic manipulation. However, existing garment reconstruction methods typically rely on unstructured representations, such as 3D Gaussian Splats, struggling to provide accurate reconstructions of garment topology and sewing structures. As a result, the reconstructed outputs are often unsuitable for high-fidelity physical simulation. We propose ReWeaver, a novel framework for topology-accurate 3D garment and sewing pattern reconstruction from sparse multi-view RGB images. Given as few as four input views, ReWeaver predicts seams and panels as well as their connectivities in both the 2D UV space and the 3D space. The predicted seams and panels align precisely with the multi-view images, yielding structured 2D--3D garment representations suitable for 3D perception, high-fidelity physical simulation, and robotic manipulation. To enable effective training, we construct a large-scale dataset GCD-TS, comprising multi-view RGB images, 3D garment geometries, textured human body meshes and annotated sewing patterns. The dataset contains over 100,000 synthetic samples covering a wide range of complex geometries and topologies. Extensive experiments show that ReWeaver consistently outperforms existing methods in terms of topology accuracy, geometry alignment and seam-panel consistency.
IF-GEO: Conflict-Aware Instruction Fusion for Multi-Query Generative Engine Optimization
Jan 20, 2026Abstract:As Generative Engines revolutionize information retrieval by synthesizing direct answers from retrieved sources, ensuring source visibility becomes a significant challenge. Improving it through targeted content revisions is a practical strategy termed Generative Engine Optimization (GEO). However, optimizing a document for diverse queries presents a constrained optimization challenge where heterogeneous queries often impose conflicting and competing revision requirements under a limited content budget. To address this challenge, we propose IF-GEO, a "diverge-then-converge" framework comprising two phases: (i) mining distinct optimization preferences from representative latent queries; (ii) synthesizing a Global Revision Blueprint for guided editing by coordinating preferences via conflict-aware instruction fusion. To explicitly quantify IF-GEO's objective of cross-query stability, we introduce risk-aware stability metrics. Experiments on multi-query benchmarks demonstrate that IF-GEO achieves substantial performance gains while maintaining robustness across diverse retrieval scenarios.
Orion-RAG: Path-Aligned Hybrid Retrieval for Graphless Data
Jan 08, 2026Abstract:Retrieval-Augmented Generation (RAG) has proven effective for knowledge synthesis, yet it encounters significant challenges in practical scenarios where data is inherently discrete and fragmented. In most environments, information is distributed across isolated files like reports and logs that lack explicit links. Standard search engines process files independently, ignoring the connections between them. Furthermore, manually building Knowledge Graphs is impractical for such vast data. To bridge this gap, we present Orion-RAG. Our core insight is simple yet effective: we do not need heavy algorithms to organize this data. Instead, we use a low-complexity strategy to extract lightweight paths that naturally link related concepts. We demonstrate that this streamlined approach suffices to transform fragmented documents into semi-structured data, enabling the system to link information across different files effectively. Extensive experiments demonstrate that Orion-RAG consistently outperforms mainstream frameworks across diverse domains, supporting real-time updates and explicit Human-in-the-Loop verification with high cost-efficiency. Experiments on FinanceBench demonstrate superior precision with a 25.2% relative improvement over strong baselines.
OxygenREC: An Instruction-Following Generative Framework for E-commerce Recommendation
Dec 31, 2025Abstract:Traditional recommendation systems suffer from inconsistency in multi-stage optimization objectives. Generative Recommendation (GR) mitigates them through an end-to-end framework; however, existing methods still rely on matching mechanisms based on inductive patterns. Although responsive, they lack the ability to uncover complex user intents that require deductive reasoning based on world knowledge. Meanwhile, LLMs show strong deep reasoning capabilities, but their latency and computational costs remain challenging for industrial applications. More critically, there are performance bottlenecks in multi-scenario scalability: as shown in Figure 1, existing solutions require independent training and deployment for each scenario, leading to low resource utilization and high maintenance costs-a challenge unaddressed in GR literature. To address these, we present OxygenREC, an industrial recommendation system that leverages Fast-Slow Thinking to deliver deep reasoning with strict latency and multi-scenario requirements of real-world environments. First, we adopt a Fast-Slow Thinking architecture. Slow thinking uses a near-line LLM pipeline to synthesize Contextual Reasoning Instructions, while fast thinking employs a high-efficiency encoder-decoder backbone for real-time generation. Second, to ensure reasoning instructions effectively enhance recommendation generation, we introduce a semantic alignment mechanism with Instruction-Guided Retrieval (IGR) to filter intent-relevant historical behaviors and use a Query-to-Item (Q2I) loss for instruction-item consistency. Finally, to resolve multi-scenario scalability, we transform scenario information into controllable instructions, using unified reward mapping and Soft Adaptive Group Clip Policy Optimization (SA-GCPO) to align policies with diverse business objectives, realizing a train-once-deploy-everywhere paradigm.
Universal Battery Degradation Forecasting Driven by Foundation Model Across Diverse Chemistries and Conditions
Dec 30, 2025Abstract:Accurate forecasting of battery capacity fade is essential for the safety, reliability, and long-term efficiency of energy storage systems. However, the strong heterogeneity across cell chemistries, form factors, and operating conditions makes it difficult to build a single model that generalizes beyond its training domain. This work proposes a unified capacity forecasting framework that maintains robust performance across diverse chemistries and usage scenarios. We curate 20 public aging datasets into a large-scale corpus covering 1,704 cells and 3,961,195 charge-discharge cycle segments, spanning temperatures from $-5\,^{\circ}\mathrm{C}$ to $45\,^{\circ}\mathrm{C}$, multiple C-rates, and application-oriented profiles such as fast charging and partial cycling. On this corpus, we adopt a Time-Series Foundation Model (TSFM) backbone and apply parameter-efficient Low-Rank Adaptation (LoRA) together with physics-guided contrastive representation learning to capture shared degradation patterns. Experiments on both seen and deliberately held-out unseen datasets show that a single unified model achieves competitive or superior accuracy compared with strong per-dataset baselines, while retaining stable performance on chemistries, capacity scales, and operating conditions excluded from training. These results demonstrate the potential of TSFM-based architectures as a scalable and transferable solution for capacity degradation forecasting in real battery management systems.
FreqDINO: Frequency-Guided Adaptation for Generalized Boundary-Aware Ultrasound Image Segmentation
Dec 12, 2025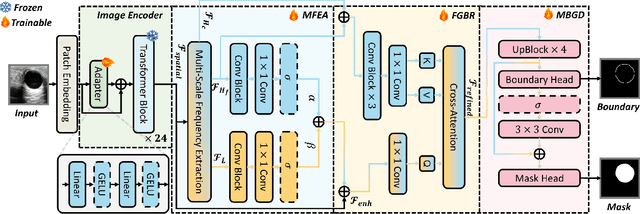
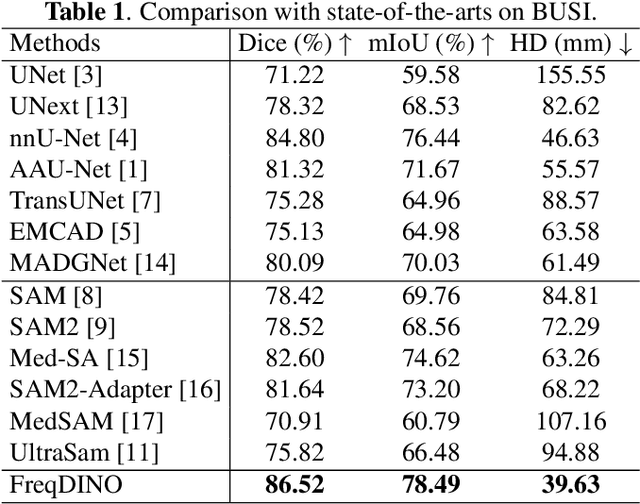
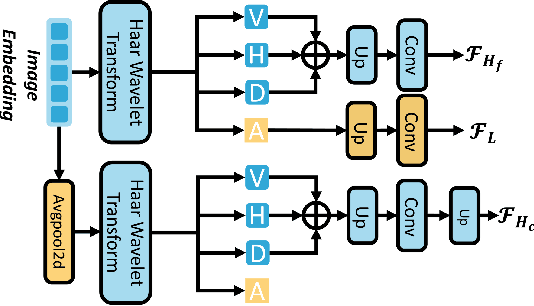
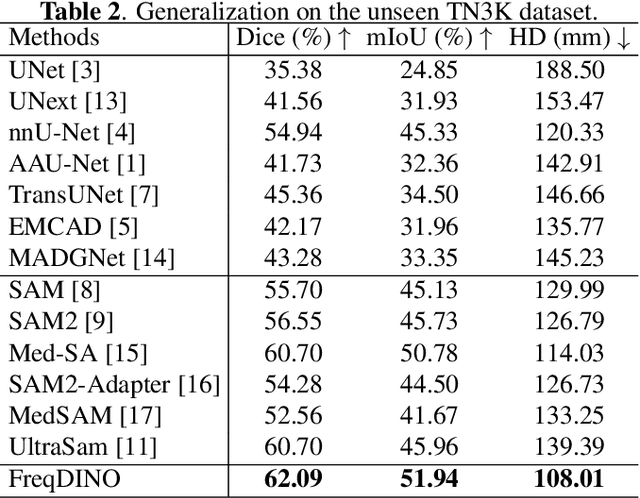
Abstract:Ultrasound image segmentation is pivotal for clinical diagnosis, yet challenged by speckle noise and imaging artifacts. Recently, DINOv3 has shown remarkable promise in medical image segmentation with its powerful representation capabilities. However, DINOv3, pre-trained on natural images, lacks sensitivity to ultrasound-specific boundary degradation. To address this limitation, we propose FreqDINO, a frequency-guided segmentation framework that enhances boundary perception and structural consistency. Specifically, we devise a Multi-scale Frequency Extraction and Alignment (MFEA) strategy to separate low-frequency structures and multi-scale high-frequency boundary details, and align them via learnable attention. We also introduce a Frequency-Guided Boundary Refinement (FGBR) module that extracts boundary prototypes from high-frequency components and refines spatial features. Furthermore, we design a Multi-task Boundary-Guided Decoder (MBGD) to ensure spatial coherence between boundary and semantic predictions. Extensive experiments demonstrate that FreqDINO surpasses state-of-the-art methods with superior achieves remarkable generalization capability. The code is at https://github.com/MingLang-FD/FreqDINO.
LapFM: A Laparoscopic Segmentation Foundation Model via Hierarchical Concept Evolving Pre-training
Dec 09, 2025Abstract:Surgical segmentation is pivotal for scene understanding yet remains hindered by annotation scarcity and semantic inconsistency across diverse procedures. Existing approaches typically fine-tune natural foundation models (e.g., SAM) with limited supervision, functioning merely as domain adapters rather than surgical foundation models. Consequently, they struggle to generalize across the vast variability of surgical targets. To bridge this gap, we present LapFM, a foundation model designed to evolve robust segmentation capabilities from massive unlabeled surgical images. Distinct from medical foundation models relying on inefficient self-supervised proxy tasks, LapFM leverages a Hierarchical Concept Evolving Pre-training paradigm. First, we establish a Laparoscopic Concept Hierarchy (LCH) via a hierarchical mask decoder with parent-child query embeddings, unifying diverse entities (i.e., Anatomy, Tissue, and Instrument) into a scalable knowledge structure with cross-granularity semantic consistency. Second, we propose a Confidence-driven Evolving Labeling that iteratively generates and filters pseudo-labels based on hierarchical consistency, progressively incorporating reliable samples from unlabeled images into training. This process yields LapBench-114K, a large-scale benchmark comprising 114K image-mask pairs. Extensive experiments demonstrate that LapFM significantly outperforms state-of-the-art methods, establishing new standards for granularity-adaptive generalization in universal laparoscopic segmentation. The source code is available at https://github.com/xq141839/LapFM.
TM-UNet: Token-Memory Enhanced Sequential Modeling for Efficient Medical Image Segmentation
Nov 15, 2025Abstract:Medical image segmentation is essential for clinical diagnosis and treatment planning. Although transformer-based methods have achieved remarkable results, their high computational cost hinders clinical deployment. To address this issue, we propose TM-UNet, a novel lightweight framework that integrates token sequence modeling with an efficient memory mechanism for efficient medical segmentation. Specifically, we introduce a multi-scale token-memory (MSTM) block that transforms 2D spatial features into token sequences through strategic spatial scanning, leveraging matrix memory cells to selectively retain and propagate discriminative contextual information across tokens. This novel token-memory mechanism acts as a dynamic knowledge store that captures long-range dependencies with linear complexity, enabling efficient global reasoning without redundant computation. Our MSTM block further incorporates exponential gating to identify token effectiveness and multi-scale contextual extraction via parallel pooling operations, enabling hierarchical representation learning without computational overhead. Extensive experiments demonstrate that TM-UNet outperforms state-of-the-art methods across diverse medical segmentation tasks with substantially reduced computation cost. The code is available at https://github.com/xq141839/TM-UNet.
Fragile by Design: On the Limits of Adversarial Defenses in Personalized Generation
Nov 13, 2025Abstract:Personalized AI applications such as DreamBooth enable the generation of customized content from user images, but also raise significant privacy concerns, particularly the risk of facial identity leakage. Recent defense mechanisms like Anti-DreamBooth attempt to mitigate this risk by injecting adversarial perturbations into user photos to prevent successful personalization. However, we identify two critical yet overlooked limitations of these methods. First, the adversarial examples often exhibit perceptible artifacts such as conspicuous patterns or stripes, making them easily detectable as manipulated content. Second, the perturbations are highly fragile, as even a simple, non-learned filter can effectively remove them, thereby restoring the model's ability to memorize and reproduce user identity. To investigate this vulnerability, we propose a novel evaluation framework, AntiDB_Purify, to systematically evaluate existing defenses under realistic purification threats, including both traditional image filters and adversarial purification. Results reveal that none of the current methods maintains their protective effectiveness under such threats. These findings highlight that current defenses offer a false sense of security and underscore the urgent need for more imperceptible and robust protections to safeguard user identity in personalized generation.
CoMA: Complementary Masking and Hierarchical Dynamic Multi-Window Self-Attention in a Unified Pre-training Framework
Nov 08, 2025Abstract:Masked Autoencoders (MAE) achieve self-supervised learning of image representations by randomly removing a portion of visual tokens and reconstructing the original image as a pretext task, thereby significantly enhancing pretraining efficiency and yielding excellent adaptability across downstream tasks. However, MAE and other MAE-style paradigms that adopt random masking generally require more pre-training epochs to maintain adaptability. Meanwhile, ViT in MAE suffers from inefficient parameter use due to fixed spatial resolution across layers. To overcome these limitations, we propose the Complementary Masked Autoencoders (CoMA), which employ a complementary masking strategy to ensure uniform sampling across all pixels, thereby improving effective learning of all features and enhancing the model's adaptability. Furthermore, we introduce DyViT, a hierarchical vision transformer that employs a Dynamic Multi-Window Self-Attention (DM-MSA), significantly reducing the parameters and FLOPs while improving fine-grained feature learning. Pre-trained on ImageNet-1K with CoMA, DyViT matches the downstream performance of MAE using only 12% of the pre-training epochs, demonstrating more effective learning. It also attains a 10% reduction in pre-training time per epoch, further underscoring its superior pre-training efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge