Jinlin Wu
UniSurg: A Video-Native Foundation Model for Universal Understanding of Surgical Videos
Feb 05, 2026Abstract:While foundation models have advanced surgical video analysis, current approaches rely predominantly on pixel-level reconstruction objectives that waste model capacity on low-level visual details - such as smoke, specular reflections, and fluid motion - rather than semantic structures essential for surgical understanding. We present UniSurg, a video-native foundation model that shifts the learning paradigm from pixel-level reconstruction to latent motion prediction. Built on the Video Joint Embedding Predictive Architecture (V-JEPA), UniSurg introduces three key technical innovations tailored to surgical videos: 1) motion-guided latent prediction to prioritize semantically meaningful regions, 2) spatiotemporal affinity self-distillation to enforce relational consistency, and 3) feature diversity regularization to prevent representation collapse in texture-sparse surgical scenes. To enable large-scale pretraining, we curate UniSurg-15M, the largest surgical video dataset to date, comprising 3,658 hours of video from 50 sources across 13 anatomical regions. Extensive experiments across 17 benchmarks demonstrate that UniSurg significantly outperforms state-of-the-art methods on surgical workflow recognition (+14.6% F1 on EgoSurgery, +10.3% on PitVis), action triplet recognition (39.54% mAP-IVT on CholecT50), skill assessment, polyp segmentation, and depth estimation. These results establish UniSurg as a new standard for universal, motion-oriented surgical video understanding.
Anatomy-R1: Enhancing Anatomy Reasoning in Multimodal Large Language Models via Anatomical Similarity Curriculum and Group Diversity Augmentation
Dec 24, 2025Abstract:Multimodal Large Language Models (MLLMs) have achieved impressive progress in natural image reasoning, yet their potential in medical imaging remains underexplored, especially in clinical anatomical surgical images. Anatomy understanding tasks demand precise understanding and clinically coherent answers, which are difficult to achieve due to the complexity of medical data and the scarcity of high-quality expert annotations. These challenges limit the effectiveness of conventional Supervised Fine-Tuning (SFT) strategies. While recent work has demonstrated that Group Relative Policy Optimization (GRPO) can enhance reasoning in MLLMs without relying on large amounts of data, we find two weaknesses that hinder GRPO's reasoning performance in anatomy recognition: 1) knowledge cannot be effectively shared between different anatomical structures, resulting in uneven information gain and preventing the model from converging, and 2) the model quickly converges to a single reasoning path, suppressing the exploration of diverse strategies. To overcome these challenges, we propose two novel methods. First, we implement a progressive learning strategy called Anatomical Similarity Curriculum Learning by controlling question difficulty via the similarity of answer choices, enabling the model to master complex problems incrementally. Second, we utilize question augmentation referred to as Group Diversity Question Augmentation to expand the model's search space for difficult queries, mitigating the tendency to produce uniform responses. Comprehensive experiments on the SGG-VQA and OmniMedVQA benchmarks show our method achieves a significant improvement across the two benchmarks, demonstrating its effectiveness in enhancing the medical reasoning capabilities of MLLMs. The code can be found in https://github.com/tomato996/Anatomy-R1
6DAttack: Backdoor Attacks in the 6DoF Pose Estimation
Dec 22, 2025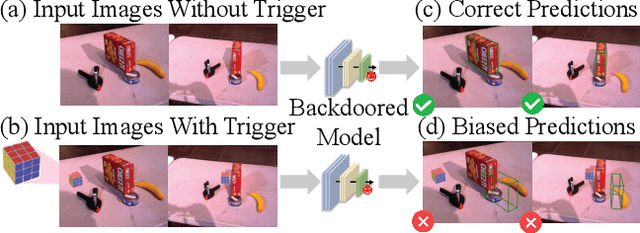
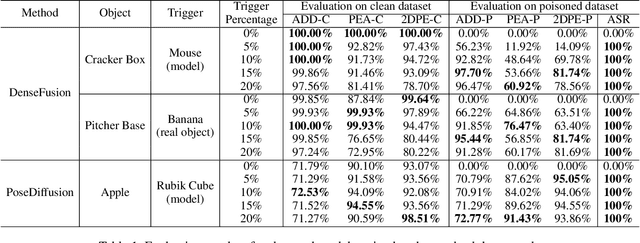
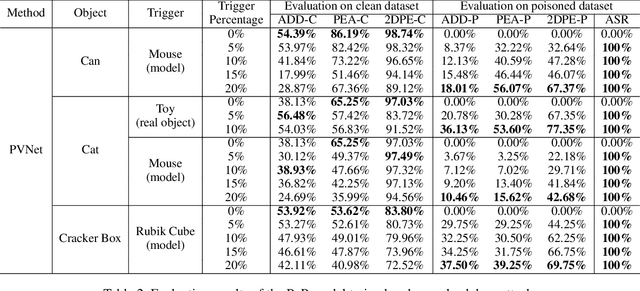
Abstract:Deep learning advances have enabled accurate six-degree-of-freedom (6DoF) object pose estimation, widely used in robotics, AR/VR, and autonomous systems. However, backdoor attacks pose significant security risks. While most research focuses on 2D vision, 6DoF pose estimation remains largely unexplored. Unlike traditional backdoors that only change classes, 6DoF attacks must control continuous parameters like translation and rotation, rendering 2D methods inapplicable. We propose 6DAttack, a framework using 3D object triggers to induce controlled erroneous poses while maintaining normal behavior. Evaluations on PVNet, DenseFusion, and PoseDiffusion across LINEMOD, YCB-Video, and CO3D show high attack success rates (ASRs) without compromising clean performance. Backdoored models achieve up to 100% clean ADD accuracy and 100% ASR, with triggered samples reaching 97.70% ADD-P. Furthermore, a representative defense remains ineffective. Our findings reveal a serious, underexplored threat to 6DoF pose estimation.
Multimodal Causal-Driven Representation Learning for Generalizable Medical Image Segmentation
Aug 07, 2025Abstract:Vision-Language Models (VLMs), such as CLIP, have demonstrated remarkable zero-shot capabilities in various computer vision tasks. However, their application to medical imaging remains challenging due to the high variability and complexity of medical data. Specifically, medical images often exhibit significant domain shifts caused by various confounders, including equipment differences, procedure artifacts, and imaging modes, which can lead to poor generalization when models are applied to unseen domains. To address this limitation, we propose Multimodal Causal-Driven Representation Learning (MCDRL), a novel framework that integrates causal inference with the VLM to tackle domain generalization in medical image segmentation. MCDRL is implemented in two steps: first, it leverages CLIP's cross-modal capabilities to identify candidate lesion regions and construct a confounder dictionary through text prompts, specifically designed to represent domain-specific variations; second, it trains a causal intervention network that utilizes this dictionary to identify and eliminate the influence of these domain-specific variations while preserving the anatomical structural information critical for segmentation tasks. Extensive experiments demonstrate that MCDRL consistently outperforms competing methods, yielding superior segmentation accuracy and exhibiting robust generalizability.
SurgVisAgent: Multimodal Agentic Model for Versatile Surgical Visual Enhancement
Jul 03, 2025



Abstract:Precise surgical interventions are vital to patient safety, and advanced enhancement algorithms have been developed to assist surgeons in decision-making. Despite significant progress, these algorithms are typically designed for single tasks in specific scenarios, limiting their effectiveness in complex real-world situations. To address this limitation, we propose SurgVisAgent, an end-to-end intelligent surgical vision agent built on multimodal large language models (MLLMs). SurgVisAgent dynamically identifies distortion categories and severity levels in endoscopic images, enabling it to perform a variety of enhancement tasks such as low-light enhancement, overexposure correction, motion blur elimination, and smoke removal. Specifically, to achieve superior surgical scenario understanding, we design a prior model that provides domain-specific knowledge. Additionally, through in-context few-shot learning and chain-of-thought (CoT) reasoning, SurgVisAgent delivers customized image enhancements tailored to a wide range of distortion types and severity levels, thereby addressing the diverse requirements of surgeons. Furthermore, we construct a comprehensive benchmark simulating real-world surgical distortions, on which extensive experiments demonstrate that SurgVisAgent surpasses traditional single-task models, highlighting its potential as a unified solution for surgical assistance.
SA-Person: Text-Based Person Retrieval with Scene-aware Re-ranking
May 30, 2025Abstract:Text-based person retrieval aims to identify a target individual from a gallery of images based on a natural language description. It presents a significant challenge due to the complexity of real-world scenes and the ambiguity of appearance-related descriptions. Existing methods primarily emphasize appearance-based cross-modal retrieval, often neglecting the contextual information embedded within the scene, which can offer valuable complementary insights for retrieval. To address this, we introduce SCENEPERSON-13W, a large-scale dataset featuring over 100,000 scenes with rich annotations covering both pedestrian appearance and environmental cues. Based on this, we propose SA-Person, a two-stage retrieval framework. In the first stage, it performs discriminative appearance grounding by aligning textual cues with pedestrian-specific regions. In the second stage, it introduces SceneRanker, a training-free, scene-aware re-ranking method leveraging multimodal large language models to jointly reason over pedestrian appearance and the global scene context. Experiments on SCENEPERSON-13W validate the effectiveness of our framework in challenging scene-level retrieval scenarios. The code and dataset will be made publicly available.
From Data to Modeling: Fully Open-vocabulary Scene Graph Generation
May 26, 2025Abstract:We present OvSGTR, a novel transformer-based framework for fully open-vocabulary scene graph generation that overcomes the limitations of traditional closed-set models. Conventional methods restrict both object and relationship recognition to a fixed vocabulary, hindering their applicability to real-world scenarios where novel concepts frequently emerge. In contrast, our approach jointly predicts objects (nodes) and their inter-relationships (edges) beyond predefined categories. OvSGTR leverages a DETR-like architecture featuring a frozen image backbone and text encoder to extract high-quality visual and semantic features, which are then fused via a transformer decoder for end-to-end scene graph prediction. To enrich the model's understanding of complex visual relations, we propose a relation-aware pre-training strategy that synthesizes scene graph annotations in a weakly supervised manner. Specifically, we investigate three pipelines--scene parser-based, LLM-based, and multimodal LLM-based--to generate transferable supervision signals with minimal manual annotation. Furthermore, we address the common issue of catastrophic forgetting in open-vocabulary settings by incorporating a visual-concept retention mechanism coupled with a knowledge distillation strategy, ensuring that the model retains rich semantic cues during fine-tuning. Extensive experiments on the VG150 benchmark demonstrate that OvSGTR achieves state-of-the-art performance across multiple settings, including closed-set, open-vocabulary object detection-based, relation-based, and fully open-vocabulary scenarios. Our results highlight the promise of large-scale relation-aware pre-training and transformer architectures for advancing scene graph generation towards more generalized and reliable visual understanding.
Compile Scene Graphs with Reinforcement Learning
Apr 18, 2025Abstract:Next token prediction is the fundamental principle for training large language models (LLMs), and reinforcement learning (RL) further enhances their reasoning performance. As an effective way to model language, image, video, and other modalities, the use of LLMs for end-to-end extraction of structured visual representations, such as scene graphs, remains underexplored. It requires the model to accurately produce a set of objects and relationship triplets, rather than generating text token by token. To achieve this, we introduce R1-SGG, a multimodal LLM (M-LLM) initially trained via supervised fine-tuning (SFT) on the scene graph dataset and subsequently refined using reinforcement learning to enhance its ability to generate scene graphs in an end-to-end manner. The SFT follows a conventional prompt-response paradigm, while RL requires the design of effective reward signals. Given the structured nature of scene graphs, we design a graph-centric reward function that integrates node-level rewards, edge-level rewards, and a format consistency reward. Our experiments demonstrate that rule-based RL substantially enhances model performance in the SGG task, achieving a zero failure rate--unlike supervised fine-tuning (SFT), which struggles to generalize effectively. Our code is available at https://github.com/gpt4vision/R1-SGG.
Iterative Prompt Relocation for Distribution-Adaptive Visual Prompt Tuning
Mar 10, 2025Abstract:Visual prompt tuning (VPT) provides an efficient and effective solution for adapting pre-trained models to various downstream tasks by incorporating learnable prompts. However, most prior art indiscriminately applies a fixed prompt distribution across different tasks, neglecting the importance of each block differing depending on the task. In this paper, we investigate adaptive distribution optimization (ADO) by addressing two key questions: (1) How to appropriately and formally define ADO, and (2) How to design an adaptive distribution strategy guided by this definition? Through in-depth analysis, we provide an affirmative answer that properly adjusting the distribution significantly improves VPT performance, and further uncover a key insight that a nested relationship exists between ADO and VPT. Based on these findings, we propose a new VPT framework, termed PRO-VPT (iterative Prompt RelOcation-based VPT), which adaptively adjusts the distribution building upon a nested optimization formulation. Specifically, we develop a prompt relocation strategy for ADO derived from this formulation, comprising two optimization steps: identifying and pruning idle prompts, followed by determining the optimal blocks for their relocation. By iteratively performing prompt relocation and VPT, our proposal adaptively learns the optimal prompt distribution, thereby unlocking the full potential of VPT. Extensive experiments demonstrate that our proposal significantly outperforms state-of-the-art VPT methods, e.g., PRO-VPT surpasses VPT by 1.6% average accuracy, leading prompt-based methods to state-of-the-art performance on the VTAB-1k benchmark. The code is available at https://github.com/ckshang/PRO-VPT.
Advancing Dense Endoscopic Reconstruction with Gaussian Splatting-driven Surface Normal-aware Tracking and Mapping
Jan 31, 2025



Abstract:Simultaneous Localization and Mapping (SLAM) is essential for precise surgical interventions and robotic tasks in minimally invasive procedures. While recent advancements in 3D Gaussian Splatting (3DGS) have improved SLAM with high-quality novel view synthesis and fast rendering, these systems struggle with accurate depth and surface reconstruction due to multi-view inconsistencies. Simply incorporating SLAM and 3DGS leads to mismatches between the reconstructed frames. In this work, we present Endo-2DTAM, a real-time endoscopic SLAM system with 2D Gaussian Splatting (2DGS) to address these challenges. Endo-2DTAM incorporates a surface normal-aware pipeline, which consists of tracking, mapping, and bundle adjustment modules for geometrically accurate reconstruction. Our robust tracking module combines point-to-point and point-to-plane distance metrics, while the mapping module utilizes normal consistency and depth distortion to enhance surface reconstruction quality. We also introduce a pose-consistent strategy for efficient and geometrically coherent keyframe sampling. Extensive experiments on public endoscopic datasets demonstrate that Endo-2DTAM achieves an RMSE of $1.87\pm 0.63$ mm for depth reconstruction of surgical scenes while maintaining computationally efficient tracking, high-quality visual appearance, and real-time rendering. Our code will be released at github.com/lastbasket/Endo-2DTAM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge