Hongbin Liu
UniSurg: A Video-Native Foundation Model for Universal Understanding of Surgical Videos
Feb 05, 2026Abstract:While foundation models have advanced surgical video analysis, current approaches rely predominantly on pixel-level reconstruction objectives that waste model capacity on low-level visual details - such as smoke, specular reflections, and fluid motion - rather than semantic structures essential for surgical understanding. We present UniSurg, a video-native foundation model that shifts the learning paradigm from pixel-level reconstruction to latent motion prediction. Built on the Video Joint Embedding Predictive Architecture (V-JEPA), UniSurg introduces three key technical innovations tailored to surgical videos: 1) motion-guided latent prediction to prioritize semantically meaningful regions, 2) spatiotemporal affinity self-distillation to enforce relational consistency, and 3) feature diversity regularization to prevent representation collapse in texture-sparse surgical scenes. To enable large-scale pretraining, we curate UniSurg-15M, the largest surgical video dataset to date, comprising 3,658 hours of video from 50 sources across 13 anatomical regions. Extensive experiments across 17 benchmarks demonstrate that UniSurg significantly outperforms state-of-the-art methods on surgical workflow recognition (+14.6% F1 on EgoSurgery, +10.3% on PitVis), action triplet recognition (39.54% mAP-IVT on CholecT50), skill assessment, polyp segmentation, and depth estimation. These results establish UniSurg as a new standard for universal, motion-oriented surgical video understanding.
LP-LLM: End-to-End Real-World Degraded License Plate Text Recognition via Large Multimodal Models
Jan 14, 2026Abstract:Real-world License Plate Recognition (LPR) faces significant challenges from severe degradations such as motion blur, low resolution, and complex illumination. The prevailing "restoration-then-recognition" two-stage paradigm suffers from a fundamental flaw: the pixel-level optimization objectives of image restoration models are misaligned with the semantic goals of character recognition, leading to artifact interference and error accumulation. While Vision-Language Models (VLMs) have demonstrated powerful general capabilities, they lack explicit structural modeling for license plate character sequences (e.g., fixed length, specific order). To address this, we propose an end-to-end structure-aware multimodal reasoning framework based on Qwen3-VL. The core innovation lies in the Character-Aware Multimodal Reasoning Module (CMRM), which introduces a set of learnable Character Slot Queries. Through a cross-attention mechanism, these queries actively retrieve fine-grained evidence corresponding to character positions from visual features. Subsequently, we inject these character-aware representations back into the visual tokens via residual modulation, enabling the language model to perform autoregressive generation based on explicit structural priors. Furthermore, combined with the LoRA parameter-efficient fine-tuning strategy, the model achieves domain adaptation while retaining the generalization capabilities of the large model. Extensive experiments on both synthetic and real-world severely degraded datasets demonstrate that our method significantly outperforms existing restoration-recognition combinations and general VLMs, validating the superiority of incorporating structured reasoning into large models for low-quality text recognition tasks.
BREATH-VL: Vision-Language-Guided 6-DoF Bronchoscopy Localization via Semantic-Geometric Fusion
Jan 07, 2026Abstract:Vision-language models (VLMs) have recently shown remarkable performance in navigation and localization tasks by leveraging large-scale pretraining for semantic understanding. However, applying VLMs to 6-DoF endoscopic camera localization presents several challenges: 1) the lack of large-scale, high-quality, densely annotated, and localization-oriented vision-language datasets in real-world medical settings; 2) limited capability for fine-grained pose regression; and 3) high computational latency when extracting temporal features from past frames. To address these issues, we first construct BREATH dataset, the largest in-vivo endoscopic localization dataset to date, collected in the complex human airway. Building on this dataset, we propose BREATH-VL, a hybrid framework that integrates semantic cues from VLMs with geometric information from vision-based registration methods for accurate 6-DoF pose estimation. Our motivation lies in the complementary strengths of both approaches: VLMs offer generalizable semantic understanding, while registration methods provide precise geometric alignment. To further enhance the VLM's ability to capture temporal context, we introduce a lightweight context-learning mechanism that encodes motion history as linguistic prompts, enabling efficient temporal reasoning without expensive video-level computation. Extensive experiments demonstrate that the vision-language module delivers robust semantic localization in challenging surgical scenes. Building on this, our BREATH-VL outperforms state-of-the-art vision-only localization methods in both accuracy and generalization, reducing translational error by 25.5% compared with the best-performing baseline, while achieving competitive computational latency.
SAMP-HDRL: Segmented Allocation with Momentum-Adjusted Utility for Multi-agent Portfolio Management via Hierarchical Deep Reinforcement Learning
Dec 28, 2025Abstract:Portfolio optimization in non-stationary markets is challenging due to regime shifts, dynamic correlations, and the limited interpretability of deep reinforcement learning (DRL) policies. We propose a Segmented Allocation with Momentum-Adjusted Utility for Multi-agent Portfolio Management via Hierarchical Deep Reinforcement Learning (SAMP-HDRL). The framework first applies dynamic asset grouping to partition the market into high-quality and ordinary subsets. An upper-level agent extracts global market signals, while lower-level agents perform intra-group allocation under mask constraints. A utility-based capital allocation mechanism integrates risky and risk-free assets, ensuring coherent coordination between global and local decisions. backtests across three market regimes (2019--2021) demonstrate that SAMP-HDRL consistently outperforms nine traditional baselines and nine DRL benchmarks under volatile and oscillating conditions. Compared with the strongest baseline, our method achieves at least 5\% higher Return, 5\% higher Sharpe ratio, 5\% higher Sortino ratio, and 2\% higher Omega ratio, with substantially larger gains observed in turbulent markets. Ablation studies confirm that upper--lower coordination, dynamic clustering, and capital allocation are indispensable to robustness. SHAP-based interpretability further reveals a complementary ``diversified + concentrated'' mechanism across agents, providing transparent insights into decision-making. Overall, SAMP-HDRL embeds structural market constraints directly into the DRL pipeline, offering improved adaptability, robustness, and interpretability in complex financial environments.
CellMamba: Adaptive Mamba for Accurate and Efficient Cell Detection
Dec 25, 2025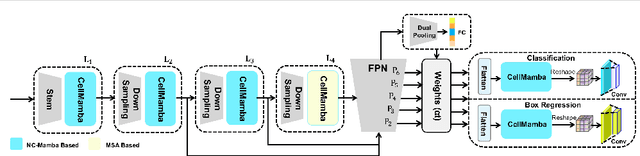
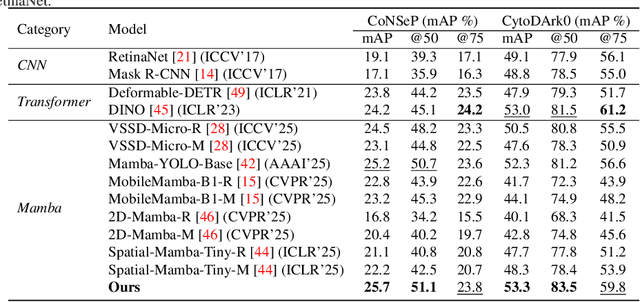
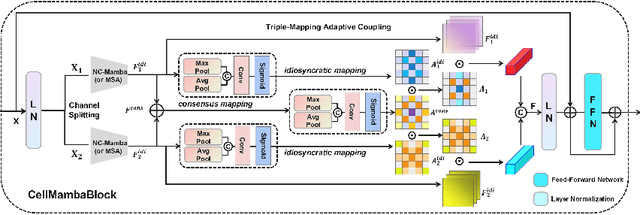
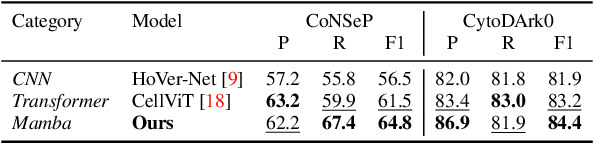
Abstract:Cell detection in pathological images presents unique challenges due to densely packed objects, subtle inter-class differences, and severe background clutter. In this paper, we propose CellMamba, a lightweight and accurate one-stage detector tailored for fine-grained biomedical instance detection. Built upon a VSSD backbone, CellMamba integrates CellMamba Blocks, which couple either NC-Mamba or Multi-Head Self-Attention (MSA) with a novel Triple-Mapping Adaptive Coupling (TMAC) module. TMAC enhances spatial discriminability by splitting channels into two parallel branches, equipped with dual idiosyncratic and one consensus attention map, adaptively fused to preserve local sensitivity and global consistency. Furthermore, we design an Adaptive Mamba Head that fuses multi-scale features via learnable weights for robust detection under varying object sizes. Extensive experiments on two public datasets-CoNSeP and CytoDArk0-demonstrate that CellMamba outperforms both CNN-based, Transformer-based, and Mamba-based baselines in accuracy, while significantly reducing model size and inference latency. Our results validate CellMamba as an efficient and effective solution for high-resolution cell detection.
Anatomy-R1: Enhancing Anatomy Reasoning in Multimodal Large Language Models via Anatomical Similarity Curriculum and Group Diversity Augmentation
Dec 24, 2025Abstract:Multimodal Large Language Models (MLLMs) have achieved impressive progress in natural image reasoning, yet their potential in medical imaging remains underexplored, especially in clinical anatomical surgical images. Anatomy understanding tasks demand precise understanding and clinically coherent answers, which are difficult to achieve due to the complexity of medical data and the scarcity of high-quality expert annotations. These challenges limit the effectiveness of conventional Supervised Fine-Tuning (SFT) strategies. While recent work has demonstrated that Group Relative Policy Optimization (GRPO) can enhance reasoning in MLLMs without relying on large amounts of data, we find two weaknesses that hinder GRPO's reasoning performance in anatomy recognition: 1) knowledge cannot be effectively shared between different anatomical structures, resulting in uneven information gain and preventing the model from converging, and 2) the model quickly converges to a single reasoning path, suppressing the exploration of diverse strategies. To overcome these challenges, we propose two novel methods. First, we implement a progressive learning strategy called Anatomical Similarity Curriculum Learning by controlling question difficulty via the similarity of answer choices, enabling the model to master complex problems incrementally. Second, we utilize question augmentation referred to as Group Diversity Question Augmentation to expand the model's search space for difficult queries, mitigating the tendency to produce uniform responses. Comprehensive experiments on the SGG-VQA and OmniMedVQA benchmarks show our method achieves a significant improvement across the two benchmarks, demonstrating its effectiveness in enhancing the medical reasoning capabilities of MLLMs. The code can be found in https://github.com/tomato996/Anatomy-R1
PADReg: Physics-Aware Deformable Registration Guided by Contact Force for Ultrasound Sequences
Aug 12, 2025Abstract:Ultrasound deformable registration estimates spatial transformations between pairs of deformed ultrasound images, which is crucial for capturing biomechanical properties and enhancing diagnostic accuracy in diseases such as thyroid nodules and breast cancer. However, ultrasound deformable registration remains highly challenging, especially under large deformation. The inherently low contrast, heavy noise and ambiguous tissue boundaries in ultrasound images severely hinder reliable feature extraction and correspondence matching. Existing methods often suffer from poor anatomical alignment and lack physical interpretability. To address the problem, we propose PADReg, a physics-aware deformable registration framework guided by contact force. PADReg leverages synchronized contact force measured by robotic ultrasound systems as a physical prior to constrain the registration. Specifically, instead of directly predicting deformation fields, we first construct a pixel-wise stiffness map utilizing the multi-modal information from contact force and ultrasound images. The stiffness map is then combined with force data to estimate a dense deformation field, through a lightweight physics-aware module inspired by Hooke's law. This design enables PADReg to achieve physically plausible registration with better anatomical alignment than previous methods relying solely on image similarity. Experiments on in-vivo datasets demonstrate that it attains a HD95 of 12.90, which is 21.34\% better than state-of-the-art methods. The source code is available at https://github.com/evelynskip/PADReg.
The Docking Game: Loop Self-Play for Fast, Dynamic, and Accurate Prediction of Flexible Protein--Ligand Binding
Aug 07, 2025Abstract:Molecular docking is a crucial aspect of drug discovery, as it predicts the binding interactions between small-molecule ligands and protein pockets. However, current multi-task learning models for docking often show inferior performance in ligand docking compared to protein pocket docking. This disparity arises largely due to the distinct structural complexities of ligands and proteins. To address this issue, we propose a novel game-theoretic framework that models the protein-ligand interaction as a two-player game called the Docking Game, with the ligand docking module acting as the ligand player and the protein pocket docking module as the protein player. To solve this game, we develop a novel Loop Self-Play (LoopPlay) algorithm, which alternately trains these players through a two-level loop. In the outer loop, the players exchange predicted poses, allowing each to incorporate the other's structural predictions, which fosters mutual adaptation over multiple iterations. In the inner loop, each player dynamically refines its predictions by incorporating its own predicted ligand or pocket poses back into its model. We theoretically show the convergence of LoopPlay, ensuring stable optimization. Extensive experiments conducted on public benchmark datasets demonstrate that LoopPlay achieves approximately a 10\% improvement in predicting accurate binding modes compared to previous state-of-the-art methods. This highlights its potential to enhance the accuracy of molecular docking in drug discovery.
Multimodal Causal-Driven Representation Learning for Generalizable Medical Image Segmentation
Aug 07, 2025Abstract:Vision-Language Models (VLMs), such as CLIP, have demonstrated remarkable zero-shot capabilities in various computer vision tasks. However, their application to medical imaging remains challenging due to the high variability and complexity of medical data. Specifically, medical images often exhibit significant domain shifts caused by various confounders, including equipment differences, procedure artifacts, and imaging modes, which can lead to poor generalization when models are applied to unseen domains. To address this limitation, we propose Multimodal Causal-Driven Representation Learning (MCDRL), a novel framework that integrates causal inference with the VLM to tackle domain generalization in medical image segmentation. MCDRL is implemented in two steps: first, it leverages CLIP's cross-modal capabilities to identify candidate lesion regions and construct a confounder dictionary through text prompts, specifically designed to represent domain-specific variations; second, it trains a causal intervention network that utilizes this dictionary to identify and eliminate the influence of these domain-specific variations while preserving the anatomical structural information critical for segmentation tasks. Extensive experiments demonstrate that MCDRL consistently outperforms competing methods, yielding superior segmentation accuracy and exhibiting robust generalizability.
MM2CT: MR-to-CT translation for multi-modal image fusion with mamba
Aug 07, 2025Abstract:Magnetic resonance (MR)-to-computed tomography (CT) translation offers significant advantages, including the elimination of radiation exposure associated with CT scans and the mitigation of imaging artifacts caused by patient motion. The existing approaches are based on single-modality MR-to-CT translation, with limited research exploring multimodal fusion. To address this limitation, we introduce Multi-modal MR to CT (MM2CT) translation method by leveraging multimodal T1- and T2-weighted MRI data, an innovative Mamba-based framework for multi-modal medical image synthesis. Mamba effectively overcomes the limited local receptive field in CNNs and the high computational complexity issues in Transformers. MM2CT leverages this advantage to maintain long-range dependencies modeling capabilities while achieving multi-modal MR feature integration. Additionally, we incorporate a dynamic local convolution module and a dynamic enhancement module to improve MRI-to-CT synthesis. The experiments on a public pelvis dataset demonstrate that MM2CT achieves state-of-the-art performance in terms of Structural Similarity Index Measure (SSIM) and Peak Signal-to-Noise Ratio (PSNR). Our code is publicly available at https://github.com/Gots-ch/MM2CT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge