Chenxin Li
MedSAM-Agent: Empowering Interactive Medical Image Segmentation with Multi-turn Agentic Reinforcement Learning
Feb 03, 2026Abstract:Medical image segmentation is evolving from task-specific models toward generalizable frameworks. Recent research leverages Multi-modal Large Language Models (MLLMs) as autonomous agents, employing reinforcement learning with verifiable reward (RLVR) to orchestrate specialized tools like the Segment Anything Model (SAM). However, these approaches often rely on single-turn, rigid interaction strategies and lack process-level supervision during training, which hinders their ability to fully exploit the dynamic potential of interactive tools and leads to redundant actions. To bridge this gap, we propose MedSAM-Agent, a framework that reformulates interactive segmentation as a multi-step autonomous decision-making process. First, we introduce a hybrid prompting strategy for expert-curated trajectory generation, enabling the model to internalize human-like decision heuristics and adaptive refinement strategies. Furthermore, we develop a two-stage training pipeline that integrates multi-turn, end-to-end outcome verification with a clinical-fidelity process reward design to promote interaction parsimony and decision efficiency. Extensive experiments across 6 medical modalities and 21 datasets demonstrate that MedSAM-Agent achieves state-of-the-art performance, effectively unifying autonomous medical reasoning with robust, iterative optimization. Code is available \href{https://github.com/CUHK-AIM-Group/MedSAM-Agent}{here}.
WonderFree: Enhancing Novel View Quality and Cross-View Consistency for 3D Scene Exploration
Jun 25, 2025Abstract:Interactive 3D scene generation from a single image has gained significant attention due to its potential to create immersive virtual worlds. However, a key challenge in current 3D generation methods is the limited explorability, which cannot render high-quality images during larger maneuvers beyond the original viewpoint, particularly when attempting to move forward into unseen areas. To address this challenge, we propose WonderFree, the first model that enables users to interactively generate 3D worlds with the freedom to explore from arbitrary angles and directions. Specifically, we decouple this challenge into two key subproblems: novel view quality, which addresses visual artifacts and floating issues in novel views, and cross-view consistency, which ensures spatial consistency across different viewpoints. To enhance rendering quality in novel views, we introduce WorldRestorer, a data-driven video restoration model designed to eliminate floaters and artifacts. In addition, a data collection pipeline is presented to automatically gather training data for WorldRestorer, ensuring it can handle scenes with varying styles needed for 3D scene generation. Furthermore, to improve cross-view consistency, we propose ConsistView, a multi-view joint restoration mechanism that simultaneously restores multiple perspectives while maintaining spatiotemporal coherence. Experimental results demonstrate that WonderFree not only enhances rendering quality across diverse viewpoints but also significantly improves global coherence and consistency. These improvements are confirmed by CLIP-based metrics and a user study showing a 77.20% preference for WonderFree over WonderWorld enabling a seamless and immersive 3D exploration experience. The code, model, and data will be publicly available.
Towards a general-purpose foundation model for fMRI analysis
Jun 11, 2025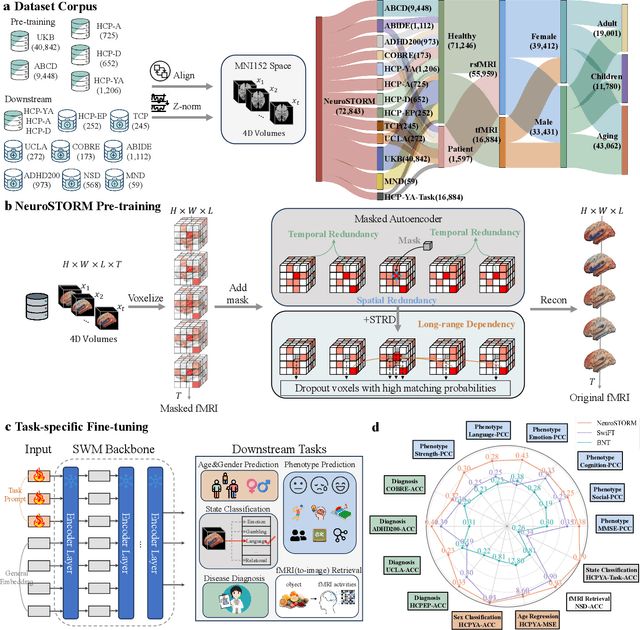

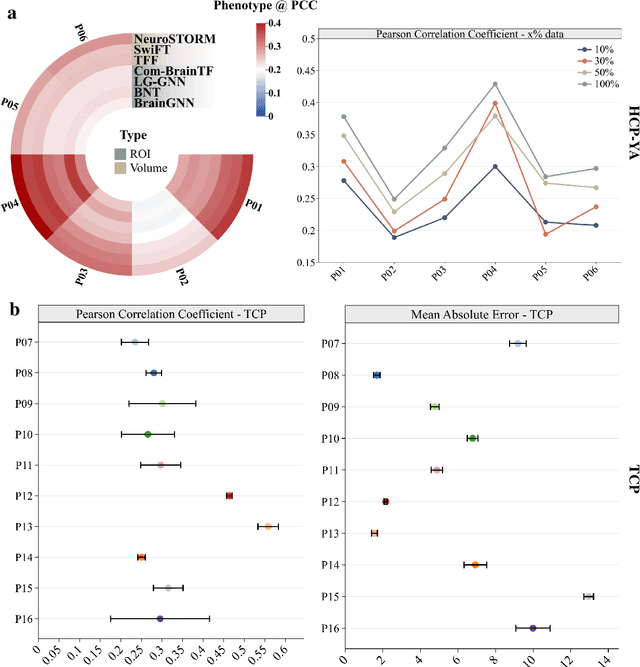
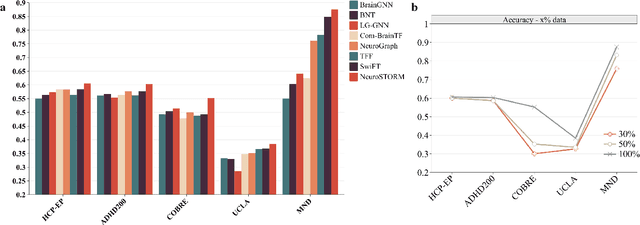
Abstract:Functional Magnetic Resonance Imaging (fMRI) is essential for studying brain function and diagnosing neurological disorders, but current analysis methods face reproducibility and transferability issues due to complex pre-processing and task-specific models. We introduce NeuroSTORM (Neuroimaging Foundation Model with Spatial-Temporal Optimized Representation Modeling), a generalizable framework that directly learns from 4D fMRI volumes and enables efficient knowledge transfer across diverse applications. NeuroSTORM is pre-trained on 28.65 million fMRI frames (>9,000 hours) from over 50,000 subjects across multiple centers and ages 5 to 100. Using a Mamba backbone and a shifted scanning strategy, it efficiently processes full 4D volumes. We also propose a spatial-temporal optimized pre-training approach and task-specific prompt tuning to improve transferability. NeuroSTORM outperforms existing methods across five tasks: age/gender prediction, phenotype prediction, disease diagnosis, fMRI-to-image retrieval, and task-based fMRI classification. It demonstrates strong clinical utility on datasets from hospitals in the U.S., South Korea, and Australia, achieving top performance in disease diagnosis and cognitive phenotype prediction. NeuroSTORM provides a standardized, open-source foundation model to improve reproducibility and transferability in fMRI-based clinical research.
Track Any Anomalous Object: A Granular Video Anomaly Detection Pipeline
Jun 05, 2025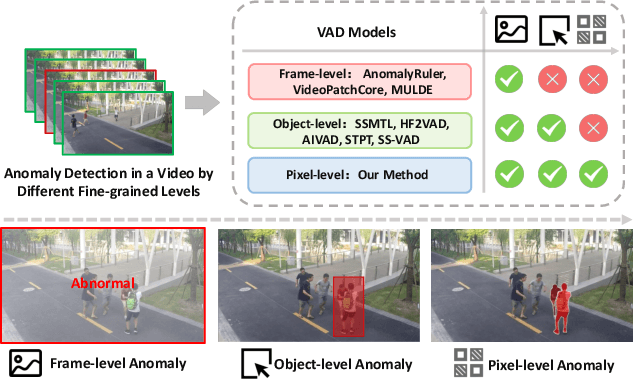
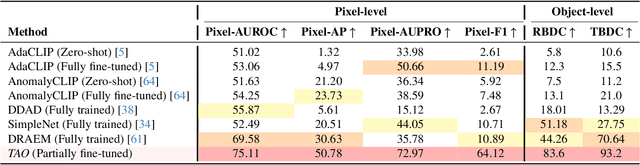
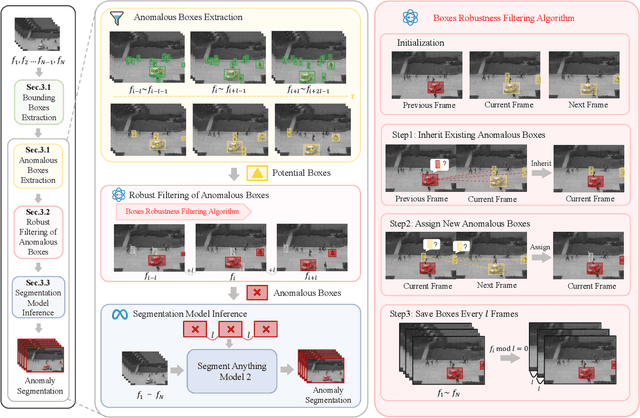

Abstract:Video anomaly detection (VAD) is crucial in scenarios such as surveillance and autonomous driving, where timely detection of unexpected activities is essential. Although existing methods have primarily focused on detecting anomalous objects in videos -- either by identifying anomalous frames or objects -- they often neglect finer-grained analysis, such as anomalous pixels, which limits their ability to capture a broader range of anomalies. To address this challenge, we propose a new framework called Track Any Anomalous Object (TAO), which introduces a granular video anomaly detection pipeline that, for the first time, integrates the detection of multiple fine-grained anomalous objects into a unified framework. Unlike methods that assign anomaly scores to every pixel, our approach transforms the problem into pixel-level tracking of anomalous objects. By linking anomaly scores to downstream tasks such as segmentation and tracking, our method removes the need for threshold tuning and achieves more precise anomaly localization in long and complex video sequences. Experiments demonstrate that TAO sets new benchmarks in accuracy and robustness. Project page available online.
FlexGS: Train Once, Deploy Everywhere with Many-in-One Flexible 3D Gaussian Splatting
Jun 04, 2025Abstract:3D Gaussian splatting (3DGS) has enabled various applications in 3D scene representation and novel view synthesis due to its efficient rendering capabilities. However, 3DGS demands relatively significant GPU memory, limiting its use on devices with restricted computational resources. Previous approaches have focused on pruning less important Gaussians, effectively compressing 3DGS but often requiring a fine-tuning stage and lacking adaptability for the specific memory needs of different devices. In this work, we present an elastic inference method for 3DGS. Given an input for the desired model size, our method selects and transforms a subset of Gaussians, achieving substantial rendering performance without additional fine-tuning. We introduce a tiny learnable module that controls Gaussian selection based on the input percentage, along with a transformation module that adjusts the selected Gaussians to complement the performance of the reduced model. Comprehensive experiments on ZipNeRF, MipNeRF and Tanks\&Temples scenes demonstrate the effectiveness of our approach. Code is available at https://flexgs.github.io.
PCMamba: Physics-Informed Cross-Modal State Space Model for Dual-Camera Compressive Hyperspectral Imaging
May 22, 2025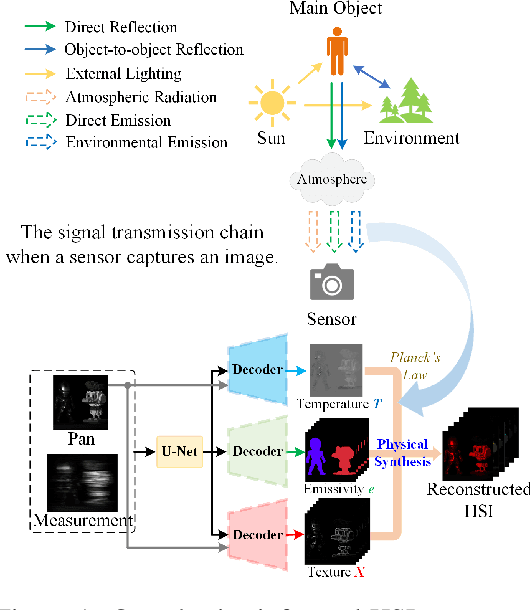
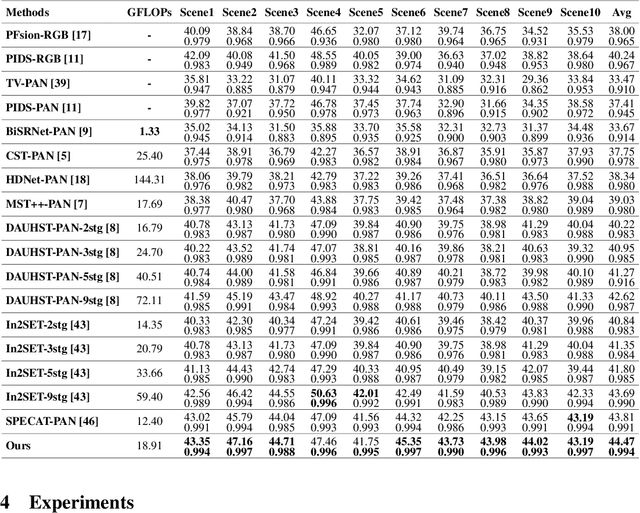
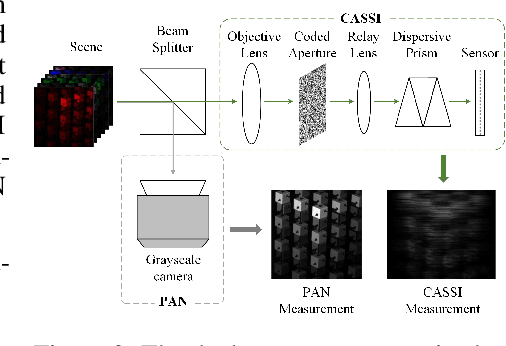
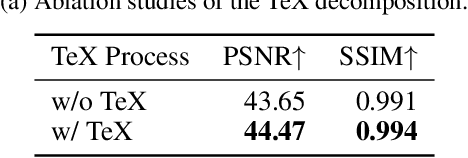
Abstract:Panchromatic (PAN) -assisted Dual-Camera Compressive Hyperspectral Imaging (DCCHI) is a key technology in snapshot hyperspectral imaging. Existing research primarily focuses on exploring spectral information from 2D compressive measurements and spatial information from PAN images in an explicit manner, leading to a bottleneck in HSI reconstruction. Various physical factors, such as temperature, emissivity, and multiple reflections between objects, play a critical role in the process of a sensor acquiring hyperspectral thermal signals. Inspired by this, we attempt to investigate the interrelationships between physical properties to provide deeper theoretical insights for HSI reconstruction. In this paper, we propose a Physics-Informed Cross-Modal State Space Model Network (PCMamba) for DCCHI, which incorporates the forward physical imaging process of HSI into the linear complexity of Mamba to facilitate lightweight and high-quality HSI reconstruction. Specifically, we analyze the imaging process of hyperspectral thermal signals to enable the network to disentangle the three key physical properties-temperature, emissivity, and texture. By fully exploiting the potential information embedded in 2D measurements and PAN images, the HSIs are reconstructed through a physics-driven synthesis process. Furthermore, we design a Cross-Modal Scanning Mamba Block (CSMB) that introduces inter-modal pixel-wise interaction with positional inductive bias by cross-scanning the backbone features and PAN features. Extensive experiments conducted on both real and simulated datasets demonstrate that our method significantly outperforms SOTA methods in both quantitative and qualitative metrics.
MonoSplat: Generalizable 3D Gaussian Splatting from Monocular Depth Foundation Models
May 21, 2025Abstract:Recent advances in generalizable 3D Gaussian Splatting have demonstrated promising results in real-time high-fidelity rendering without per-scene optimization, yet existing approaches still struggle to handle unfamiliar visual content during inference on novel scenes due to limited generalizability. To address this challenge, we introduce MonoSplat, a novel framework that leverages rich visual priors from pre-trained monocular depth foundation models for robust Gaussian reconstruction. Our approach consists of two key components: a Mono-Multi Feature Adapter that transforms monocular features into multi-view representations, coupled with an Integrated Gaussian Prediction module that effectively fuses both feature types for precise Gaussian generation. Through the Adapter's lightweight attention mechanism, features are seamlessly aligned and aggregated across views while preserving valuable monocular priors, enabling the Prediction module to generate Gaussian primitives with accurate geometry and appearance. Through extensive experiments on diverse real-world datasets, we convincingly demonstrate that MonoSplat achieves superior reconstruction quality and generalization capability compared to existing methods while maintaining computational efficiency with minimal trainable parameters. Codes are available at https://github.com/CUHK-AIM-Group/MonoSplat.
X-GRM: Large Gaussian Reconstruction Model for Sparse-view X-rays to Computed Tomography
May 21, 2025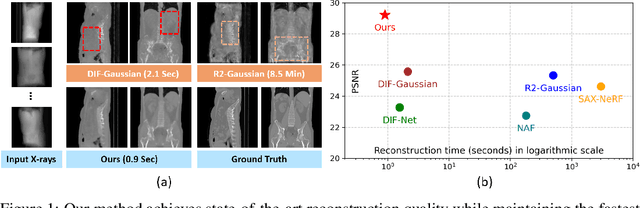
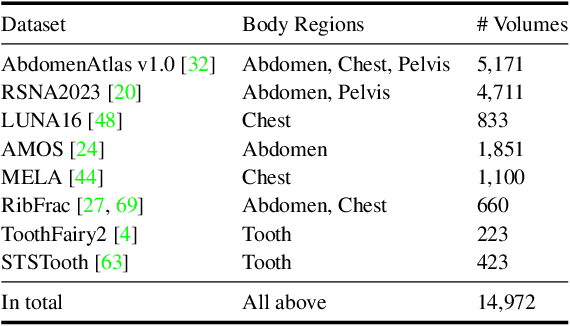
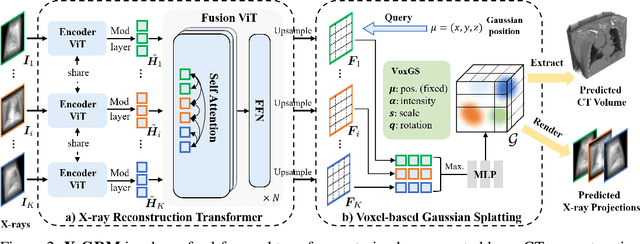
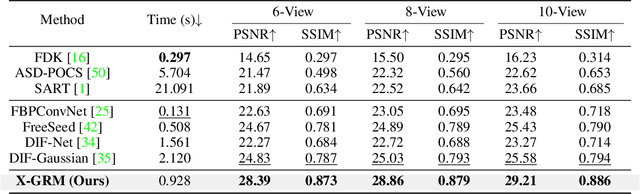
Abstract:Computed Tomography serves as an indispensable tool in clinical workflows, providing non-invasive visualization of internal anatomical structures. Existing CT reconstruction works are limited to small-capacity model architecture, inflexible volume representation, and small-scale training data. In this paper, we present X-GRM (X-ray Gaussian Reconstruction Model), a large feedforward model for reconstructing 3D CT from sparse-view 2D X-ray projections. X-GRM employs a scalable transformer-based architecture to encode an arbitrary number of sparse X-ray inputs, where tokens from different views are integrated efficiently. Then, tokens are decoded into a new volume representation, named Voxel-based Gaussian Splatting (VoxGS), which enables efficient CT volume extraction and differentiable X-ray rendering. To support the training of X-GRM, we collect ReconX-15K, a large-scale CT reconstruction dataset containing around 15,000 CT/X-ray pairs across diverse organs, including the chest, abdomen, pelvis, and tooth etc. This combination of a high-capacity model, flexible volume representation, and large-scale training data empowers our model to produce high-quality reconstructions from various testing inputs, including in-domain and out-domain X-ray projections. Project Page: https://github.com/CUHK-AIM-Group/X-GRM.
HRMedSeg: Unlocking High-resolution Medical Image segmentation via Memory-efficient Attention Modeling
Apr 08, 2025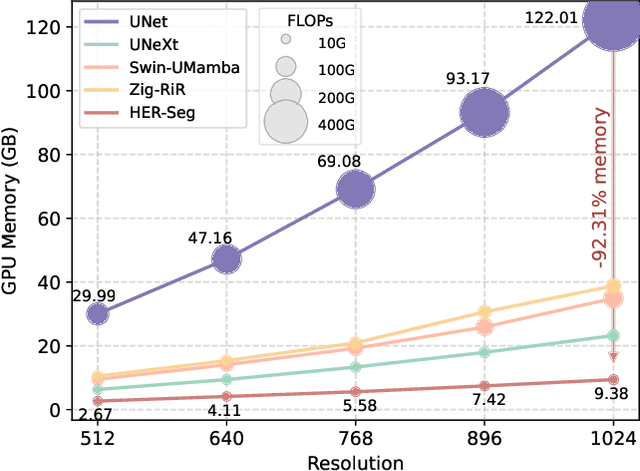
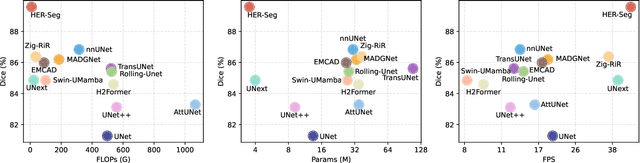
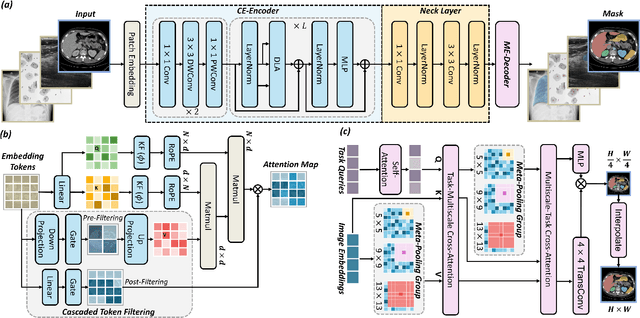
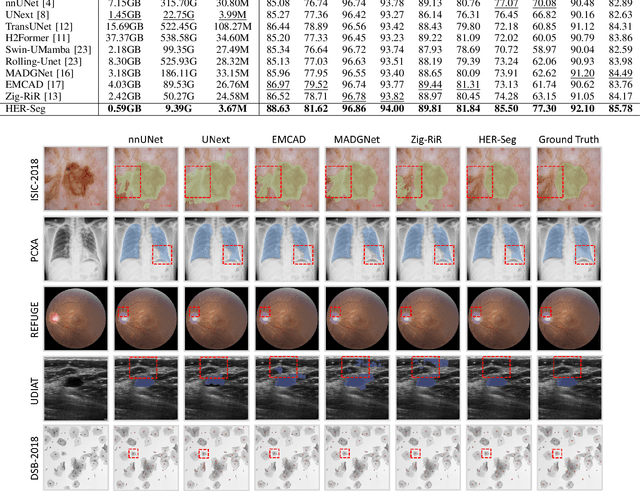
Abstract:High-resolution segmentation is critical for precise disease diagnosis by extracting micro-imaging information from medical images. Existing transformer-based encoder-decoder frameworks have demonstrated remarkable versatility and zero-shot performance in medical segmentation. While beneficial, they usually require huge memory costs when handling large-size segmentation mask predictions, which are expensive to apply to real-world scenarios. To address this limitation, we propose a memory-efficient framework for high-resolution medical image segmentation, called HRMedSeg. Specifically, we first devise a lightweight gated vision transformer (LGViT) as our image encoder to model long-range dependencies with linear complexity. Then, we design an efficient cross-multiscale decoder (ECM-Decoder) to generate high-resolution segmentation masks. Moreover, we utilize feature distillation during pretraining to unleash the potential of our proposed model. Extensive experiments reveal that HRMedSeg outperforms state-of-the-arts in diverse high-resolution medical image segmentation tasks. In particular, HRMedSeg uses only 0.59GB GPU memory per batch during fine-tuning, demonstrating low training costs. Besides, when HRMedSeg meets the Segment Anything Model (SAM), our HRMedSegSAM takes 0.61% parameters of SAM-H. The code is available at https://github.com/xq141839/HRMedSeg.
JarvisIR: Elevating Autonomous Driving Perception with Intelligent Image Restoration
Apr 05, 2025



Abstract:Vision-centric perception systems struggle with unpredictable and coupled weather degradations in the wild. Current solutions are often limited, as they either depend on specific degradation priors or suffer from significant domain gaps. To enable robust and autonomous operation in real-world conditions, we propose JarvisIR, a VLM-powered agent that leverages the VLM as a controller to manage multiple expert restoration models. To further enhance system robustness, reduce hallucinations, and improve generalizability in real-world adverse weather, JarvisIR employs a novel two-stage framework consisting of supervised fine-tuning and human feedback alignment. Specifically, to address the lack of paired data in real-world scenarios, the human feedback alignment enables the VLM to be fine-tuned effectively on large-scale real-world data in an unsupervised manner. To support the training and evaluation of JarvisIR, we introduce CleanBench, a comprehensive dataset consisting of high-quality and large-scale instruction-responses pairs, including 150K synthetic entries and 80K real entries. Extensive experiments demonstrate that JarvisIR exhibits superior decision-making and restoration capabilities. Compared with existing methods, it achieves a 50% improvement in the average of all perception metrics on CleanBench-Real. Project page: https://cvpr2025-jarvisir.github.io/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge