Yupei Zhang
Joint Modelling Histology and Molecular Markers for Cancer Classification
Feb 11, 2025
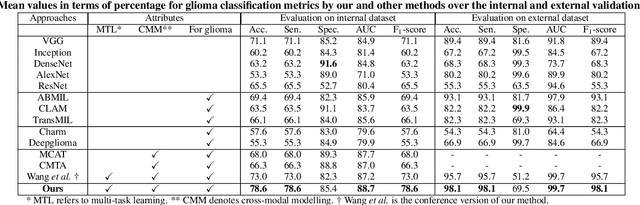
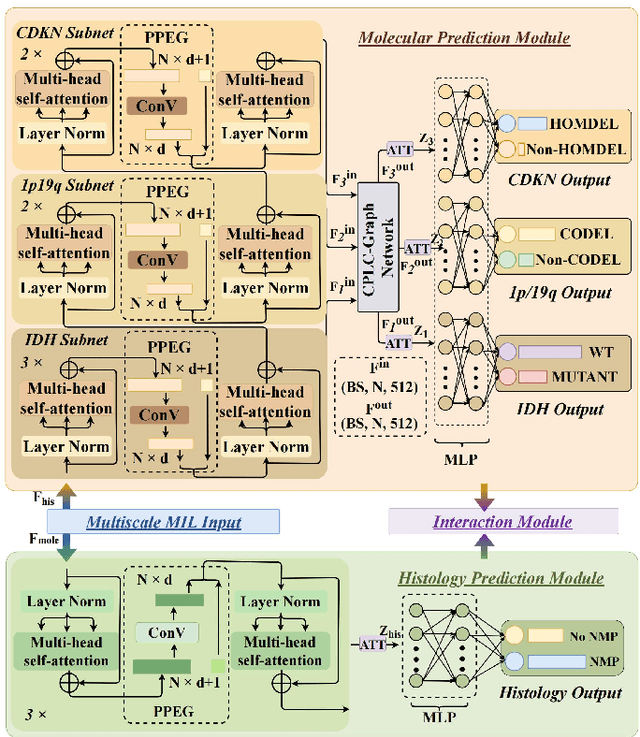
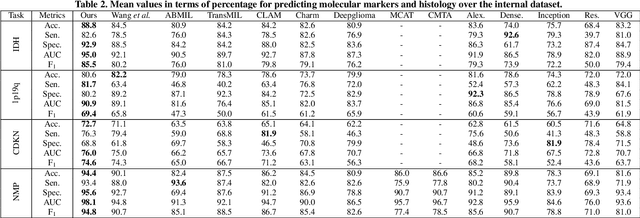
Abstract:Cancers are characterized by remarkable heterogeneity and diverse prognosis. Accurate cancer classification is essential for patient stratification and clinical decision-making. Although digital pathology has been advancing cancer diagnosis and prognosis, the paradigm in cancer pathology has shifted from purely relying on histology features to incorporating molecular markers. There is an urgent need for digital pathology methods to meet the needs of the new paradigm. We introduce a novel digital pathology approach to jointly predict molecular markers and histology features and model their interactions for cancer classification. Firstly, to mitigate the challenge of cross-magnification information propagation, we propose a multi-scale disentangling module, enabling the extraction of multi-scale features from high-magnification (cellular-level) to low-magnification (tissue-level) whole slide images. Further, based on the multi-scale features, we propose an attention-based hierarchical multi-task multi-instance learning framework to simultaneously predict histology and molecular markers. Moreover, we propose a co-occurrence probability-based label correlation graph network to model the co-occurrence of molecular markers. Lastly, we design a cross-modal interaction module with the dynamic confidence constrain loss and a cross-modal gradient modulation strategy, to model the interactions of histology and molecular markers. Our experiments demonstrate that our method outperforms other state-of-the-art methods in classifying glioma, histology features and molecular markers. Our method promises to promote precise oncology with the potential to advance biomedical research and clinical applications. The code is available at https://github.com/LHY1007/M3C2
Long-tailed Medical Diagnosis with Relation-aware Representation Learning and Iterative Classifier Calibration
Feb 05, 2025



Abstract:Recently computer-aided diagnosis has demonstrated promising performance, effectively alleviating the workload of clinicians. However, the inherent sample imbalance among different diseases leads algorithms biased to the majority categories, leading to poor performance for rare categories. Existing works formulated this challenge as a long-tailed problem and attempted to tackle it by decoupling the feature representation and classification. Yet, due to the imbalanced distribution and limited samples from tail classes, these works are prone to biased representation learning and insufficient classifier calibration. To tackle these problems, we propose a new Long-tailed Medical Diagnosis (LMD) framework for balanced medical image classification on long-tailed datasets. In the initial stage, we develop a Relation-aware Representation Learning (RRL) scheme to boost the representation ability by encouraging the encoder to capture intrinsic semantic features through different data augmentations. In the subsequent stage, we propose an Iterative Classifier Calibration (ICC) scheme to calibrate the classifier iteratively. This is achieved by generating a large number of balanced virtual features and fine-tuning the encoder using an Expectation-Maximization manner. The proposed ICC compensates for minority categories to facilitate unbiased classifier optimization while maintaining the diagnostic knowledge in majority classes. Comprehensive experiments on three public long-tailed medical datasets demonstrate that our LMD framework significantly surpasses state-of-the-art approaches. The source code can be accessed at https://github.com/peterlipan/LMD.
Federated Deep Subspace Clustering
Dec 31, 2024Abstract:This paper introduces FDSC, a private-protected subspace clustering (SC) approach with federated learning (FC) schema. In each client, there is a deep subspace clustering network accounting for grouping the isolated data, composed of a encode network, a self-expressive layer, and a decode network. FDSC is achieved by uploading the encode network to communicate with other clients in the server. Besides, FDSC is also enhanced by preserving the local neighborhood relationship in each client. With the effects of federated learning and locality preservation, the learned data features from the encoder are boosted so as to enhance the self-expressiveness learning and result in better clustering performance. Experiments test FDSC on public datasets and compare with other clustering methods, demonstrating the effectiveness of FDSC.
Cooperative SQL Generation for Segmented Databases By Using Multi-functional LLM Agents
Dec 08, 2024



Abstract:Text-to-SQL task aims to automatically yield SQL queries according to user text questions. To address this problem, we propose a Cooperative SQL Generation framework based on Multi-functional Agents (CSMA) through information interaction among large language model (LLM) based agents who own part of the database schema seperately. Inspired by the collaboration in human teamwork, CSMA consists of three stages: 1) Question-related schema collection, 2) Question-corresponding SQL query generation, and 3) SQL query correctness check. In the first stage, agents analyze their respective schema and communicate with each other to collect the schema information relevant to the question. In the second stage, agents try to generate the corresponding SQL query for the question using the collected information. In the third stage, agents check if the SQL query is created correctly according to their known information. This interaction-based method makes the question-relevant part of database schema from each agent to be used for SQL generation and check. Experiments on the Spider and Bird benckmark demonstrate that CSMA achieves a high performance level comparable to the state-of-the-arts, meanwhile holding the private data in these individual agents.
Focus on Focus: Focus-oriented Representation Learning and Multi-view Cross-modal Alignment for Glioma Grading
Aug 16, 2024



Abstract:Recently, multimodal deep learning, which integrates histopathology slides and molecular biomarkers, has achieved a promising performance in glioma grading. Despite great progress, due to the intra-modality complexity and inter-modality heterogeneity, existing studies suffer from inadequate histopathology representation learning and inefficient molecular-pathology knowledge alignment. These two issues hinder existing methods to precisely interpret diagnostic molecular-pathology features, thereby limiting their grading performance. Moreover, the real-world applicability of existing multimodal approaches is significantly restricted as molecular biomarkers are not always available during clinical deployment. To address these problems, we introduce a novel Focus on Focus (FoF) framework with paired pathology-genomic training and applicable pathology-only inference, enhancing molecular-pathology representation effectively. Specifically, we propose a Focus-oriented Representation Learning (FRL) module to encourage the model to identify regions positively or negatively related to glioma grading and guide it to focus on the diagnostic areas with a consistency constraint. To effectively link the molecular biomarkers to morphological features, we propose a Multi-view Cross-modal Alignment (MCA) module that projects histopathology representations into molecular subspaces, aligning morphological features with corresponding molecular biomarker status by supervised contrastive learning. Experiments on the TCGA GBM-LGG dataset demonstrate that our FoF framework significantly improves the glioma grading. Remarkably, our FoF achieves superior performance using only histopathology slides compared to existing multimodal methods. The source code is available at https://github.com/peterlipan/FoF.
Knowledge-driven Subspace Fusion and Gradient Coordination for Multi-modal Learning
Jun 20, 2024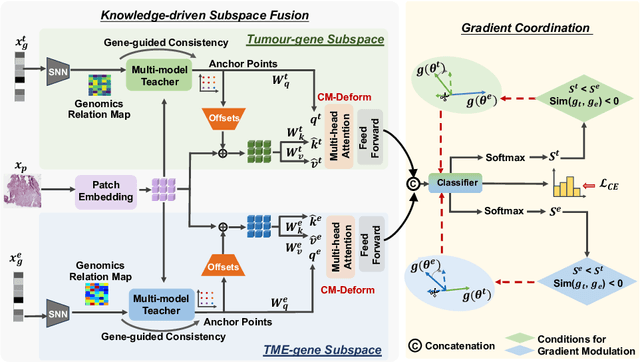
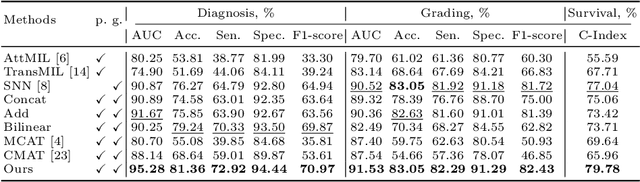

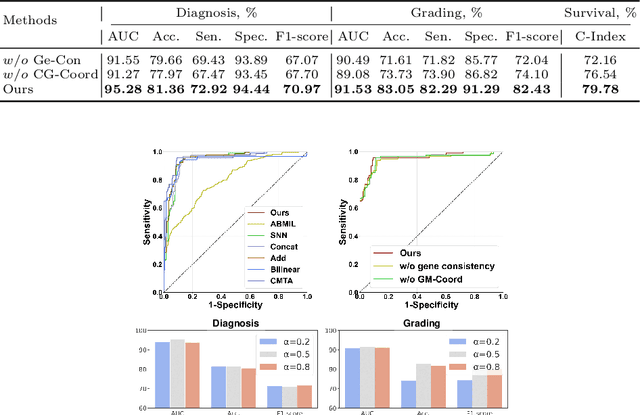
Abstract:Multi-modal learning plays a crucial role in cancer diagnosis and prognosis. Current deep learning based multi-modal approaches are often limited by their abilities to model the complex correlations between genomics and histology data, addressing the intrinsic complexity of tumour ecosystem where both tumour and microenvironment contribute to malignancy. We propose a biologically interpretative and robust multi-modal learning framework to efficiently integrate histology images and genomics by decomposing the feature subspace of histology images and genomics, reflecting distinct tumour and microenvironment features. To enhance cross-modal interactions, we design a knowledge-driven subspace fusion scheme, consisting of a cross-modal deformable attention module and a gene-guided consistency strategy. Additionally, in pursuit of dynamically optimizing the subspace knowledge, we further propose a novel gradient coordination learning strategy. Extensive experiments demonstrate the effectiveness of the proposed method, outperforming state-of-the-art techniques in three downstream tasks of glioma diagnosis, tumour grading, and survival analysis. Our code is available at https://github.com/helenypzhang/Subspace-Multimodal-Learning.
Unified Multi-modal Diagnostic Framework with Reconstruction Pre-training and Heterogeneity-combat Tuning
Apr 09, 2024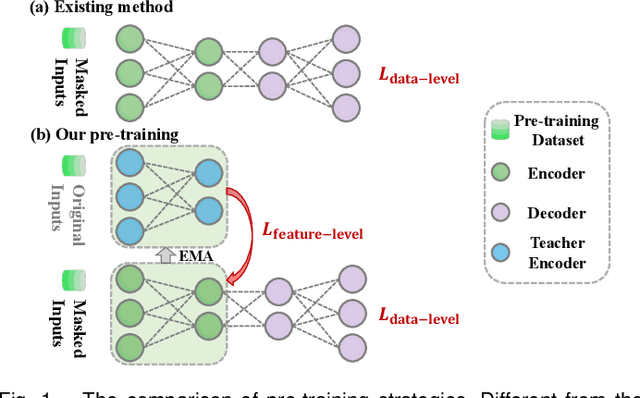
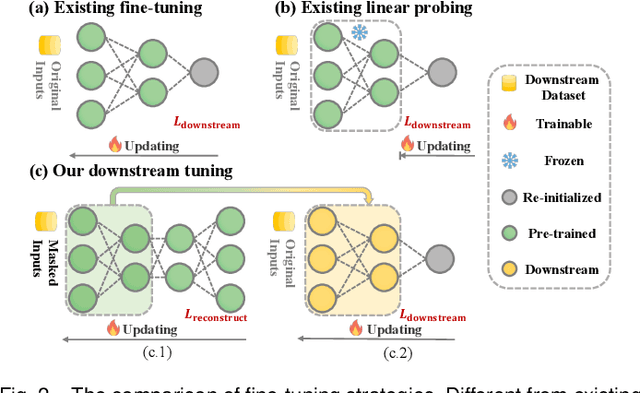
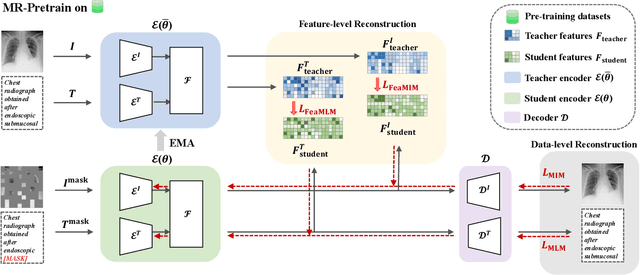
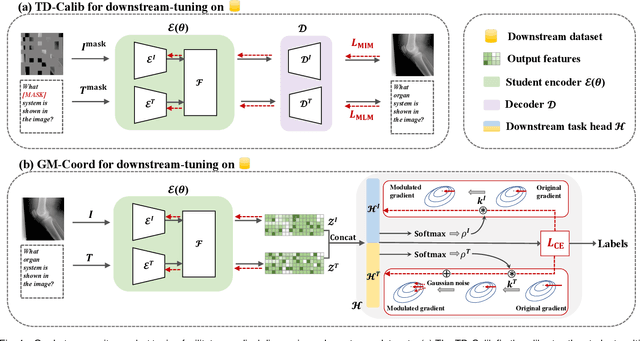
Abstract:Medical multi-modal pre-training has revealed promise in computer-aided diagnosis by leveraging large-scale unlabeled datasets. However, existing methods based on masked autoencoders mainly rely on data-level reconstruction tasks, but lack high-level semantic information. Furthermore, two significant heterogeneity challenges hinder the transfer of pre-trained knowledge to downstream tasks, \textit{i.e.}, the distribution heterogeneity between pre-training data and downstream data, and the modality heterogeneity within downstream data. To address these challenges, we propose a Unified Medical Multi-modal Diagnostic (UMD) framework with tailored pre-training and downstream tuning strategies. Specifically, to enhance the representation abilities of vision and language encoders, we propose the Multi-level Reconstruction Pre-training (MR-Pretrain) strategy, including a feature-level and data-level reconstruction, which guides models to capture the semantic information from masked inputs of different modalities. Moreover, to tackle two kinds of heterogeneities during the downstream tuning, we present the heterogeneity-combat downstream tuning strategy, which consists of a Task-oriented Distribution Calibration (TD-Calib) and a Gradient-guided Modality Coordination (GM-Coord). In particular, TD-Calib fine-tunes the pre-trained model regarding the distribution of downstream datasets, and GM-Coord adjusts the gradient weights according to the dynamic optimization status of different modalities. Extensive experiments on five public medical datasets demonstrate the effectiveness of our UMD framework, which remarkably outperforms existing approaches on three kinds of downstream tasks.
Federated learning-outcome prediction with multi-layer privacy protection
Dec 25, 2023Abstract:Learning-outcome prediction (LOP) is a long-standing and critical problem in educational routes. Many studies have contributed to developing effective models while often suffering from data shortage and low generalization to various institutions due to the privacy-protection issue. To this end, this study proposes a distributed grade prediction model, dubbed FecMap, by exploiting the federated learning (FL) framework that preserves the private data of local clients and communicates with others through a global generalized model. FecMap considers local subspace learning (LSL), which explicitly learns the local features against the global features, and multi-layer privacy protection (MPP), which hierarchically protects the private features, including model-shareable features and not-allowably shared features, to achieve client-specific classifiers of high performance on LOP per institution. FecMap is then achieved in an iteration manner with all datasets distributed on clients by training a local neural network composed of a global part, a local part, and a classification head in clients and averaging the global parts from clients on the server. To evaluate the FecMap model, we collected three higher-educational datasets of student academic records from engineering majors. Experiment results manifest that FecMap benefits from the proposed LSL and MPP and achieves steady performance on the task of LOP, compared with the state-of-the-art models. This study makes a fresh attempt at the use of federated learning in the learning-analytical task, potentially paving the way to facilitating personalized education with privacy protection.
* 10 pages, 9 figures, 3 tables. This preprint will be published in Frontiers of Computer Science on Dec 15, 2024
Concept Prerequisite Relation Prediction by Using Permutation-Equivariant Directed Graph Neural Networks
Dec 15, 2023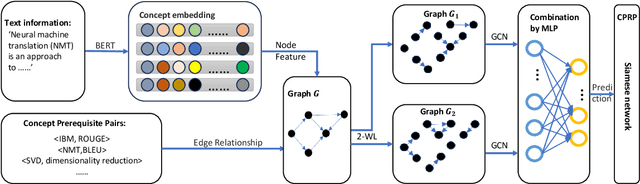
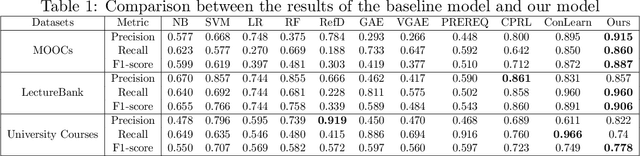
Abstract:This paper studies the problem of CPRP, concept prerequisite relation prediction, which is a fundamental task in using AI for education. CPRP is usually formulated into a link-prediction task on a relationship graph of concepts and solved by training the graph neural network (GNN) model. However, current directed GNNs fail to manage graph isomorphism which refers to the invariance of non-isomorphic graphs, reducing the expressivity of resulting representations. We present a permutation-equivariant directed GNN model by introducing the Weisfeiler-Lehman test into directed GNN learning. Our method is then used for CPRP and evaluated on three public datasets. The experimental results show that our model delivers better prediction performance than the state-of-the-art methods.
Landmark Tracking in Liver US images Using Cascade Convolutional Neural Networks with Long Short-Term Memory
Sep 14, 2022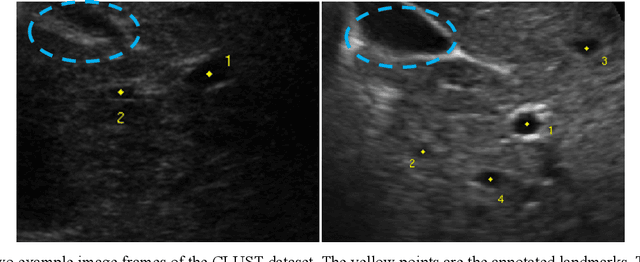
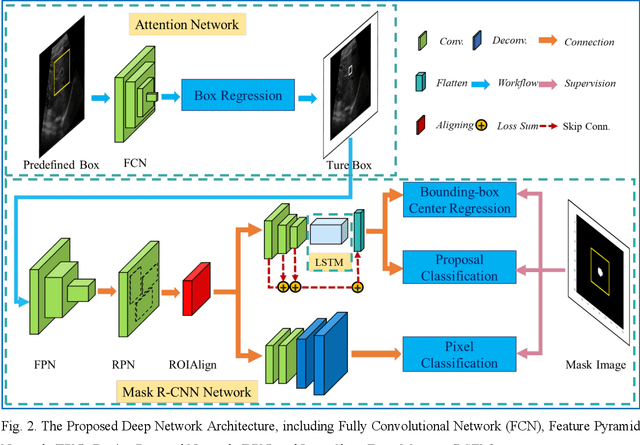
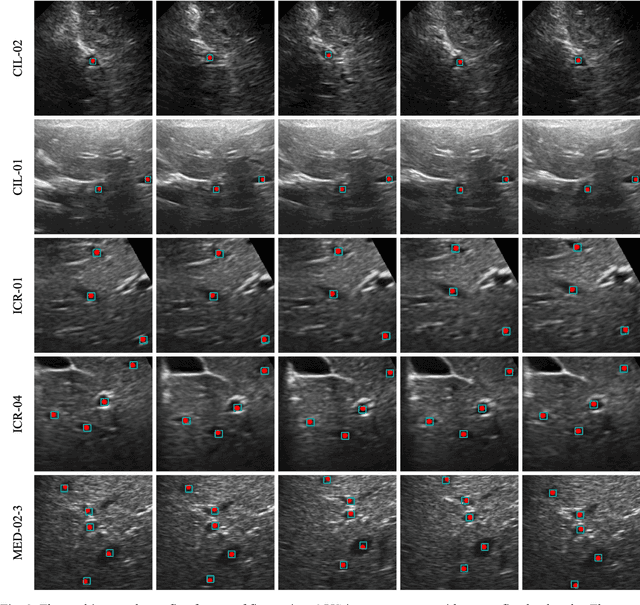
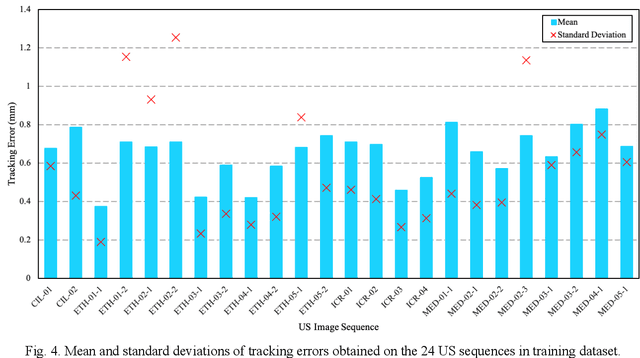
Abstract:This study proposed a deep learning-based tracking method for ultrasound (US) image-guided radiation therapy. The proposed cascade deep learning model is composed of an attention network, a mask region-based convolutional neural network (mask R-CNN), and a long short-term memory (LSTM) network. The attention network learns a mapping from a US image to a suspected area of landmark motion in order to reduce the search region. The mask R-CNN then produces multiple region-of-interest (ROI) proposals in the reduced region and identifies the proposed landmark via three network heads: bounding box regression, proposal classification, and landmark segmentation. The LSTM network models the temporal relationship among the successive image frames for bounding box regression and proposal classification. To consolidate the final proposal, a selection method is designed according to the similarities between sequential frames. The proposed method was tested on the liver US tracking datasets used in the Medical Image Computing and Computer Assisted Interventions (MICCAI) 2015 challenges, where the landmarks were annotated by three experienced observers to obtain their mean positions. Five-fold cross-validation on the 24 given US sequences with ground truths shows that the mean tracking error for all landmarks is 0.65+/-0.56 mm, and the errors of all landmarks are within 2 mm. We further tested the proposed model on 69 landmarks from the testing dataset that has a similar image pattern to the training pattern, resulting in a mean tracking error of 0.94+/-0.83 mm. Our experimental results have demonstrated the feasibility and accuracy of our proposed method in tracking liver anatomic landmarks using US images, providing a potential solution for real-time liver tracking for active motion management during radiation therapy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge