Hao Duan
Understanding Learning Invariance in Deep Linear Networks
Jun 16, 2025Abstract:Equivariant and invariant machine learning models exploit symmetries and structural patterns in data to improve sample efficiency. While empirical studies suggest that data-driven methods such as regularization and data augmentation can perform comparably to explicitly invariant models, theoretical insights remain scarce. In this paper, we provide a theoretical comparison of three approaches for achieving invariance: data augmentation, regularization, and hard-wiring. We focus on mean squared error regression with deep linear networks, which parametrize rank-bounded linear maps and can be hard-wired to be invariant to specific group actions. We show that the critical points of the optimization problems for hard-wiring and data augmentation are identical, consisting solely of saddles and the global optimum. By contrast, regularization introduces additional critical points, though they remain saddles except for the global optimum. Moreover, we demonstrate that the regularization path is continuous and converges to the hard-wired solution.
Joint Modelling Histology and Molecular Markers for Cancer Classification
Feb 11, 2025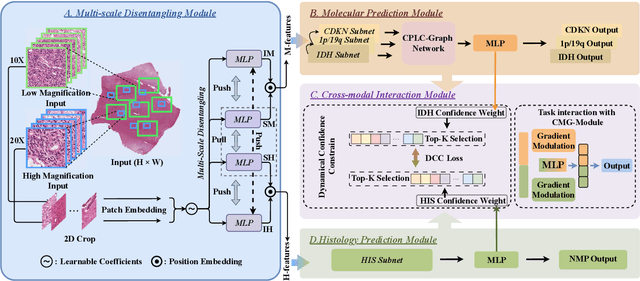
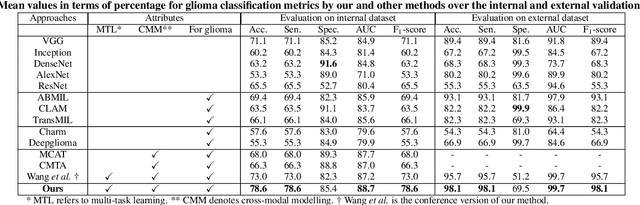
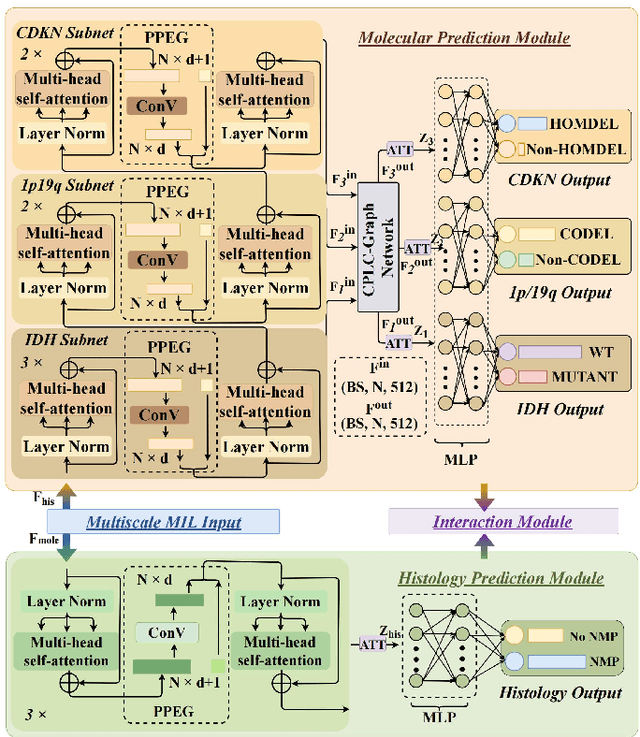
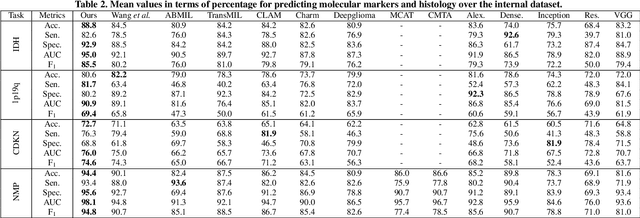
Abstract:Cancers are characterized by remarkable heterogeneity and diverse prognosis. Accurate cancer classification is essential for patient stratification and clinical decision-making. Although digital pathology has been advancing cancer diagnosis and prognosis, the paradigm in cancer pathology has shifted from purely relying on histology features to incorporating molecular markers. There is an urgent need for digital pathology methods to meet the needs of the new paradigm. We introduce a novel digital pathology approach to jointly predict molecular markers and histology features and model their interactions for cancer classification. Firstly, to mitigate the challenge of cross-magnification information propagation, we propose a multi-scale disentangling module, enabling the extraction of multi-scale features from high-magnification (cellular-level) to low-magnification (tissue-level) whole slide images. Further, based on the multi-scale features, we propose an attention-based hierarchical multi-task multi-instance learning framework to simultaneously predict histology and molecular markers. Moreover, we propose a co-occurrence probability-based label correlation graph network to model the co-occurrence of molecular markers. Lastly, we design a cross-modal interaction module with the dynamic confidence constrain loss and a cross-modal gradient modulation strategy, to model the interactions of histology and molecular markers. Our experiments demonstrate that our method outperforms other state-of-the-art methods in classifying glioma, histology features and molecular markers. Our method promises to promote precise oncology with the potential to advance biomedical research and clinical applications. The code is available at https://github.com/LHY1007/M3C2
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge