Stephen Price
Latent-Constrained Conditional VAEs for Augmenting Large-Scale Climate Ensembles
Jan 01, 2026Abstract:Large climate-model ensembles are computationally expensive; yet many downstream analyses would benefit from additional, statistically consistent realizations of spatiotemporal climate variables. We study a generative modeling approach for producing new realizations from a limited set of available runs by transferring structure learned across an ensemble. Using monthly near-surface temperature time series from ten independent reanalysis realizations (ERA5), we find that a vanilla conditional variational autoencoder (CVAE) trained jointly across realizations yields a fragmented latent space that fails to generalize to unseen ensemble members. To address this, we introduce a latent-constrained CVAE (LC-CVAE) that enforces cross-realization homogeneity of latent embeddings at a small set of shared geographic 'anchor' locations. We then use multi-output Gaussian process regression in the latent space to predict latent coordinates at unsampled locations in a new realization, followed by decoding to generate full time series fields. Experiments and ablations demonstrate (i) instability when training on a single realization, (ii) diminishing returns after incorporating roughly five realizations, and (iii) a trade-off between spatial coverage and reconstruction quality that is closely linked to the average neighbor distance in latent space.
Joint Modelling Histology and Molecular Markers for Cancer Classification
Feb 11, 2025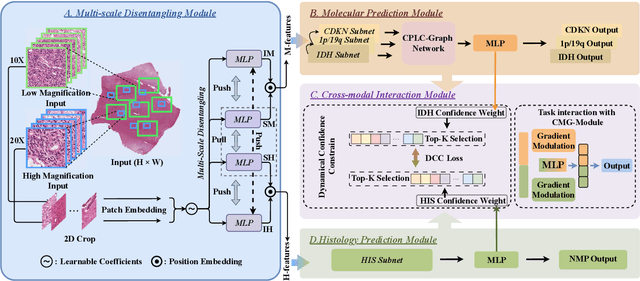
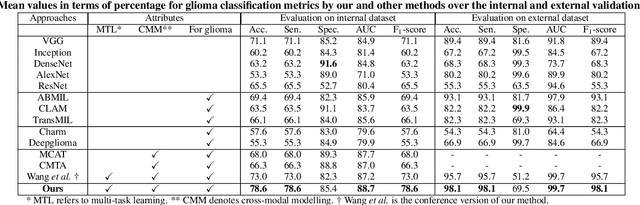
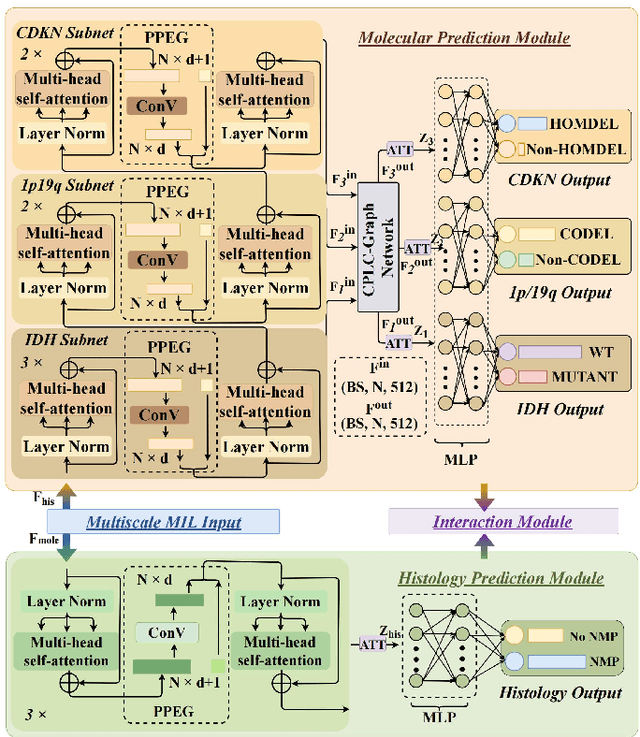
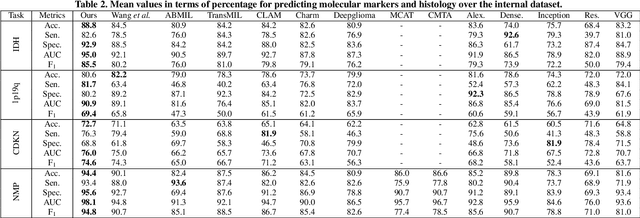
Abstract:Cancers are characterized by remarkable heterogeneity and diverse prognosis. Accurate cancer classification is essential for patient stratification and clinical decision-making. Although digital pathology has been advancing cancer diagnosis and prognosis, the paradigm in cancer pathology has shifted from purely relying on histology features to incorporating molecular markers. There is an urgent need for digital pathology methods to meet the needs of the new paradigm. We introduce a novel digital pathology approach to jointly predict molecular markers and histology features and model their interactions for cancer classification. Firstly, to mitigate the challenge of cross-magnification information propagation, we propose a multi-scale disentangling module, enabling the extraction of multi-scale features from high-magnification (cellular-level) to low-magnification (tissue-level) whole slide images. Further, based on the multi-scale features, we propose an attention-based hierarchical multi-task multi-instance learning framework to simultaneously predict histology and molecular markers. Moreover, we propose a co-occurrence probability-based label correlation graph network to model the co-occurrence of molecular markers. Lastly, we design a cross-modal interaction module with the dynamic confidence constrain loss and a cross-modal gradient modulation strategy, to model the interactions of histology and molecular markers. Our experiments demonstrate that our method outperforms other state-of-the-art methods in classifying glioma, histology features and molecular markers. Our method promises to promote precise oncology with the potential to advance biomedical research and clinical applications. The code is available at https://github.com/LHY1007/M3C2
Multi-task Learning of Histology and Molecular Markers for Classifying Diffuse Glioma
Mar 26, 2023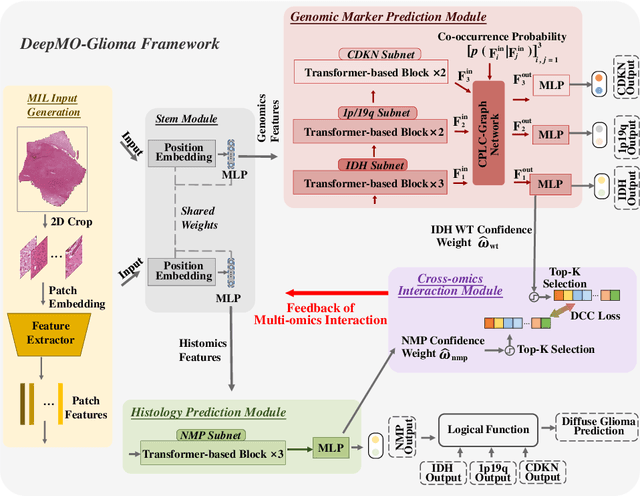

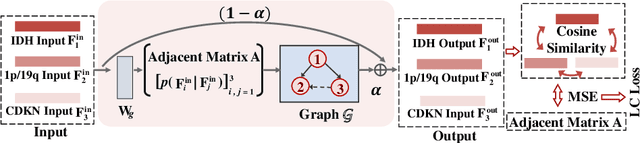

Abstract:Most recently, the pathology diagnosis of cancer is shifting to integrating molecular makers with histology features. It is a urgent need for digital pathology methods to effectively integrate molecular markers with histology, which could lead to more accurate diagnosis in the real world scenarios. This paper presents a first attempt to jointly predict molecular markers and histology features and model their interactions for classifying diffuse glioma bases on whole slide images. Specifically, we propose a hierarchical multi-task multi-instance learning framework to jointly predict histology and molecular markers. Moreover, we propose a co-occurrence probability-based label correction graph network to model the co-occurrence of molecular markers. Lastly, we design an inter-omic interaction strategy with the dynamical confidence constraint loss to model the interactions of histology and molecular markers. Our experiments show that our method outperforms other state-of-the-art methods in classifying diffuse glioma,as well as related histology and molecular markers on a multi-institutional dataset.
Mutual Contrastive Learning to Disentangle Whole Slide Image Representations for Glioma Grading
Mar 08, 2022

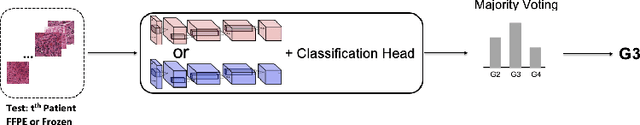
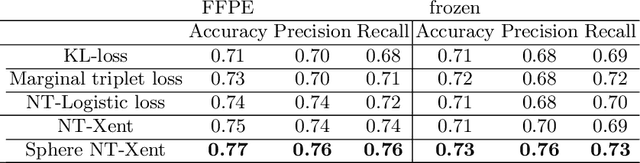
Abstract:Whole slide images (WSI) provide valuable phenotypic information for histological assessment and malignancy grading of tumors. The WSI-based computational pathology promises to provide rapid diagnostic support and facilitate digital health. The most commonly used WSI are derived from formalin-fixed paraffin-embedded (FFPE) and frozen sections. Currently, the majority of automatic tumor grading models are developed based on FFPE sections, which could be affected by the artifacts introduced by tissue processing. Here we propose a mutual contrastive learning scheme to integrate FFPE and frozen sections and disentangle cross-modality representations for glioma grading. We first design a mutual learning scheme to jointly optimize the model training based on FFPE and frozen sections. Further, we develop a multi-modality domain alignment mechanism to ensure semantic consistency in the backbone model training. We finally design a sphere normalized temperature-scaled cross-entropy loss (NT-Xent), which could promote cross-modality representation disentangling of FFPE and frozen sections. Our experiments show that the proposed scheme achieves better performance than the model trained based on each single modality or mixed modalities. The sphere NT-Xent loss outperforms other typical metrics loss functions.
Adaptive unsupervised learning with enhanced feature representation for intra-tumor partitioning and survival prediction for glioblastoma
Aug 21, 2021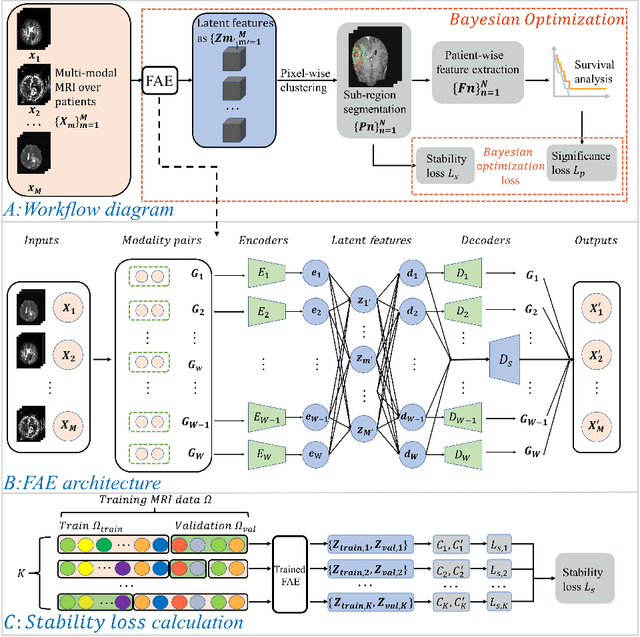
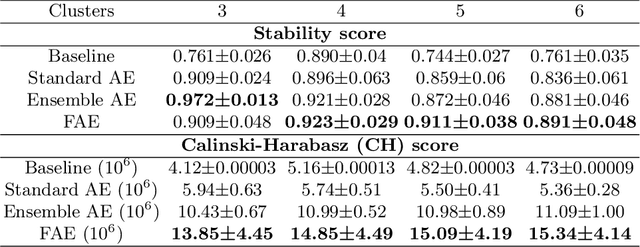
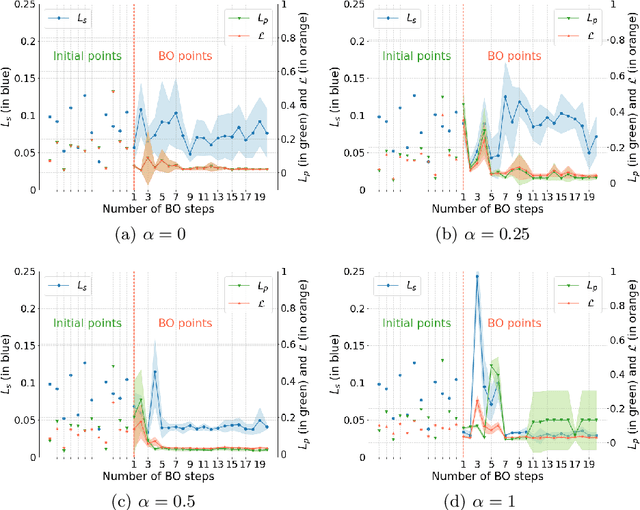
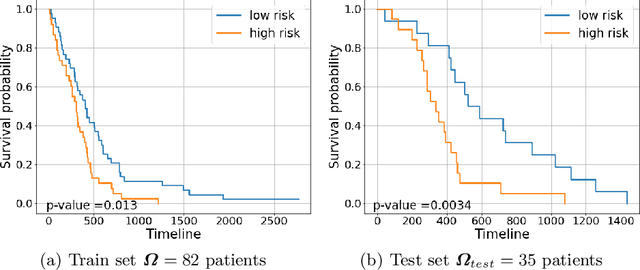
Abstract:Glioblastoma is profoundly heterogeneous in regional microstructure and vasculature. Characterizing the spatial heterogeneity of glioblastoma could lead to more precise treatment. With unsupervised learning techniques, glioblastoma MRI-derived radiomic features have been widely utilized for tumor sub-region segmentation and survival prediction. However, the reliability of algorithm outcomes is often challenged by both ambiguous intermediate process and instability introduced by the randomness of clustering algorithms, especially for data from heterogeneous patients. In this paper, we propose an adaptive unsupervised learning approach for efficient MRI intra-tumor partitioning and glioblastoma survival prediction. A novel and problem-specific Feature-enhanced Auto-Encoder (FAE) is developed to enhance the representation of pairwise clinical modalities and therefore improve clustering stability of unsupervised learning algorithms such as K-means. Moreover, the entire process is modelled by the Bayesian optimization (BO) technique with a custom loss function that the hyper-parameters can be adaptively optimized in a reasonably few steps. The results demonstrate that the proposed approach can produce robust and clinically relevant MRI sub-regions and statistically significant survival predictions.
Bayesian optimization assisted unsupervised learning for efficient intra-tumor partitioning in MRI and survival prediction for glioblastoma patients
Dec 05, 2020

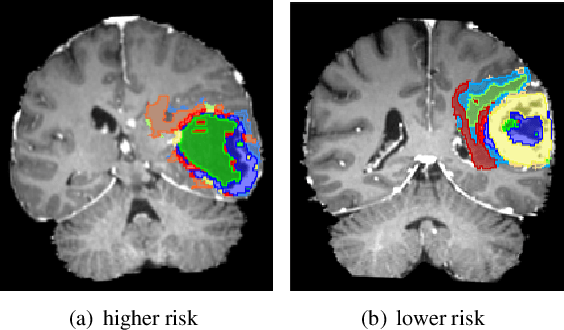
Abstract:Glioblastoma is profoundly heterogeneous in microstructure and vasculature, which may lead to tumor regional diversity and distinct treatment response. Although successful in tumor sub-region segmentation and survival prediction, radiomics based on machine learning algorithms, is challenged by its robustness, due to the vague intermediate process and track changes. Also, the weak interpretability of the model poses challenges to clinical application. Here we proposed a machine learning framework to semi-automatically fine-tune the clustering algorithms and quantitatively identify stable sub-regions for reliable clinical survival prediction. Hyper-parameters are automatically determined by the global minimum of the trained Gaussian Process (GP) surrogate model through Bayesian optimization(BO) to alleviate the difficulty of tuning parameters for clinical researchers. To enhance the interpretability of the survival prediction model, we incorporated the prior knowledge of intra-tumoral heterogeneity, by segmenting tumor sub-regions and extracting sub-regional features. The results demonstrated that the global minimum of the trained GP surrogate can be used as sub-optimal hyper-parameter solutions for efficient. The sub-regions segmented based on physiological MRI can be applied to predict patient survival, which could enhance the clinical interpretability for the machine learning model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge