Lipei Zhang
Brain Foundation Models with Hypergraph Dynamic Adapter for Brain Disease Analysis
May 01, 2025Abstract:Brain diseases, such as Alzheimer's disease and brain tumors, present profound challenges due to their complexity and societal impact. Recent advancements in brain foundation models have shown significant promise in addressing a range of brain-related tasks. However, current brain foundation models are limited by task and data homogeneity, restricted generalization beyond segmentation or classification, and inefficient adaptation to diverse clinical tasks. In this work, we propose SAM-Brain3D, a brain-specific foundation model trained on over 66,000 brain image-label pairs across 14 MRI sub-modalities, and Hypergraph Dynamic Adapter (HyDA), a lightweight adapter for efficient and effective downstream adaptation. SAM-Brain3D captures detailed brain-specific anatomical and modality priors for segmenting diverse brain targets and broader downstream tasks. HyDA leverages hypergraphs to fuse complementary multi-modal data and dynamically generate patient-specific convolutional kernels for multi-scale feature fusion and personalized patient-wise adaptation. Together, our framework excels across a broad spectrum of brain disease segmentation and classification tasks. Extensive experiments demonstrate that our method consistently outperforms existing state-of-the-art approaches, offering a new paradigm for brain disease analysis through multi-modal, multi-scale, and dynamic foundation modeling.
D2SA: Dual-Stage Distribution and Slice Adaptation for Efficient Test-Time Adaptation in MRI Reconstruction
Mar 25, 2025Abstract:Variations in Magnetic resonance imaging (MRI) scanners and acquisition protocols cause distribution shifts that degrade reconstruction performance on unseen data. Test-time adaptation (TTA) offers a promising solution to address this discrepancies. However, previous single-shot TTA approaches are inefficient due to repeated training and suboptimal distributional models. Self-supervised learning methods are also limited by scarce date scenarios. To address these challenges, we propose a novel Dual-Stage Distribution and Slice Adaptation (D2SA) via MRI implicit neural representation (MR-INR) to improve MRI reconstruction performance and efficiency, which features two stages. In the first stage, an MR-INR branch performs patient-wise distribution adaptation by learning shared representations across slices and modelling patient-specific shifts with mean and variance adjustments. In the second stage, single-slice adaptation refines the output from frozen convolutional layers with a learnable anisotropic diffusion module, preventing over-smoothing and reducing computation. Experiments across four MRI distribution shifts demonstrate that our method can integrate well with various self-supervised learning (SSL) framework, improving performance and accelerating convergence under diverse conditions.
Where Do We Stand with Implicit Neural Representations? A Technical and Performance Survey
Nov 06, 2024



Abstract:Implicit Neural Representations (INRs) have emerged as a paradigm in knowledge representation, offering exceptional flexibility and performance across a diverse range of applications. INRs leverage multilayer perceptrons (MLPs) to model data as continuous implicit functions, providing critical advantages such as resolution independence, memory efficiency, and generalisation beyond discretised data structures. Their ability to solve complex inverse problems makes them particularly effective for tasks including audio reconstruction, image representation, 3D object reconstruction, and high-dimensional data synthesis. This survey provides a comprehensive review of state-of-the-art INR methods, introducing a clear taxonomy that categorises them into four key areas: activation functions, position encoding, combined strategies, and network structure optimisation. We rigorously analyse their critical properties, such as full differentiability, smoothness, compactness, and adaptability to varying resolutions while also examining their strengths and limitations in addressing locality biases and capturing fine details. Our experimental comparison offers new insights into the trade-offs between different approaches, showcasing the capabilities and challenges of the latest INR techniques across various tasks. In addition to identifying areas where current methods excel, we highlight key limitations and potential avenues for improvement, such as developing more expressive activation functions, enhancing positional encoding mechanisms, and improving scalability for complex, high-dimensional data. This survey serves as a roadmap for researchers, offering practical guidance for future exploration in the field of INRs. We aim to foster new methodologies by outlining promising research directions for INRs and applications.
Biophysics Informed Pathological Regularisation for Brain Tumour Segmentation
Mar 18, 2024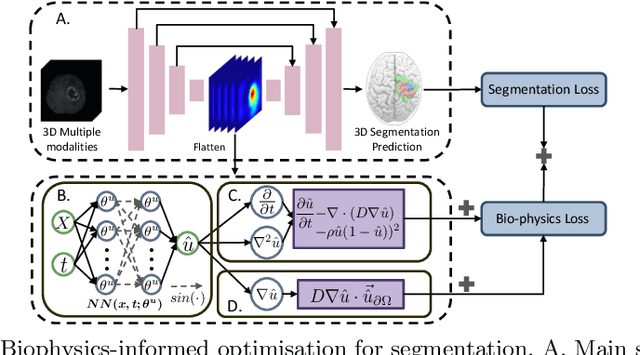
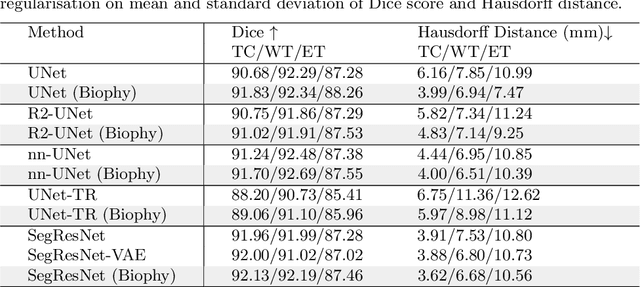
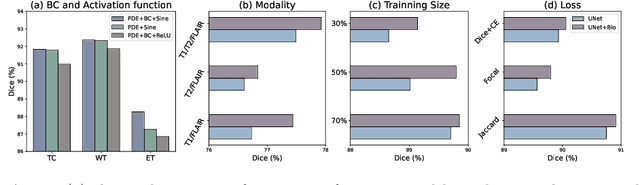
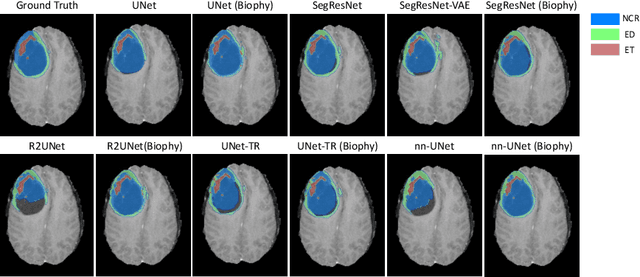
Abstract:Recent advancements in deep learning have significantly improved brain tumour segmentation techniques; however, the results still lack confidence and robustness as they solely consider image data without biophysical priors or pathological information. Integrating biophysics-informed regularisation is one effective way to change this situation, as it provides an prior regularisation for automated end-to-end learning. In this paper, we propose a novel approach that designs brain tumour growth Partial Differential Equation (PDE) models as a regularisation with deep learning, operational with any network model. Our method introduces tumour growth PDE models directly into the segmentation process, improving accuracy and robustness, especially in data-scarce scenarios. This system estimates tumour cell density using a periodic activation function. By effectively integrating this estimation with biophysical models, we achieve a better capture of tumour characteristics. This approach not only aligns the segmentation closer to actual biological behaviour but also strengthens the model's performance under limited data conditions. We demonstrate the effectiveness of our framework through extensive experiments on the BraTS 2023 dataset, showcasing significant improvements in both precision and reliability of tumour segmentation.
Single-Shot Plug-and-Play Methods for Inverse Problems
Nov 22, 2023



Abstract:The utilisation of Plug-and-Play (PnP) priors in inverse problems has become increasingly prominent in recent years. This preference is based on the mathematical equivalence between the general proximal operator and the regularised denoiser, facilitating the adaptation of various off-the-shelf denoiser priors to a wide range of inverse problems. However, existing PnP models predominantly rely on pre-trained denoisers using large datasets. In this work, we introduce Single-Shot PnP methods (SS-PnP), shifting the focus to solving inverse problems with minimal data. First, we integrate Single-Shot proximal denoisers into iterative methods, enabling training with single instances. Second, we propose implicit neural priors based on a novel function that preserves relevant frequencies to capture fine details while avoiding the issue of vanishing gradients. We demonstrate, through extensive numerical and visual experiments, that our method leads to better approximations.
Beyond U: Making Diffusion Models Faster & Lighter
Oct 31, 2023Abstract:Diffusion models are a family of generative models that yield record-breaking performance in tasks such as image synthesis, video generation, and molecule design. Despite their capabilities, their efficiency, especially in the reverse denoising process, remains a challenge due to slow convergence rates and high computational costs. In this work, we introduce an approach that leverages continuous dynamical systems to design a novel denoising network for diffusion models that is more parameter-efficient, exhibits faster convergence, and demonstrates increased noise robustness. Experimenting with denoising probabilistic diffusion models, our framework operates with approximately a quarter of the parameters and 30% of the Floating Point Operations (FLOPs) compared to standard U-Nets in Denoising Diffusion Probabilistic Models (DDPMs). Furthermore, our model is up to 70% faster in inference than the baseline models when measured in equal conditions while converging to better quality solutions.
Attention-based 3D CNN with Multi-layer Features for Alzheimer's Disease Diagnosis using Brain Images
Aug 10, 2023Abstract:Structural MRI and PET imaging play an important role in the diagnosis of Alzheimer's disease (AD), showing the morphological changes and glucose metabolism changes in the brain respectively. The manifestations in the brain image of some cognitive impairment patients are relatively inconspicuous, for example, it still has difficulties in achieving accurate diagnosis through sMRI in clinical practice. With the emergence of deep learning, convolutional neural network (CNN) has become a valuable method in AD-aided diagnosis, but some CNN methods cannot effectively learn the features of brain image, making the diagnosis of AD still presents some challenges. In this work, we propose an end-to-end 3D CNN framework for AD diagnosis based on ResNet, which integrates multi-layer features obtained under the effect of the attention mechanism to better capture subtle differences in brain images. The attention maps showed our model can focus on key brain regions related to the disease diagnosis. Our method was verified in ablation experiments with two modality images on 792 subjects from the ADNI database, where AD diagnostic accuracies of 89.71% and 91.18% were achieved based on sMRI and PET respectively, and also outperformed some state-of-the-art methods.
* 4 pages, 4 figures
Multi-modal learning for predicting the genotype of glioma
Mar 21, 2022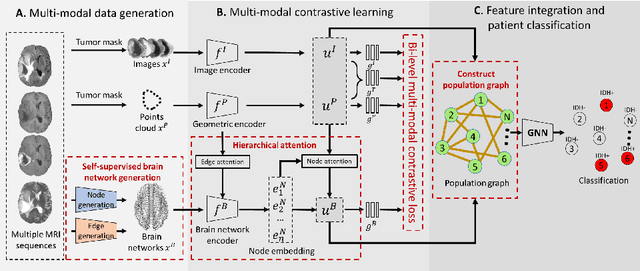
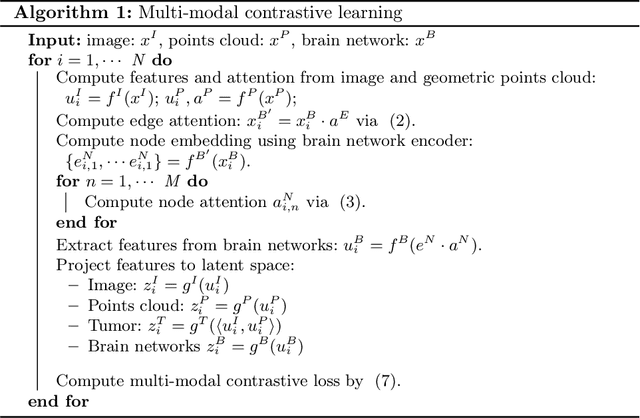
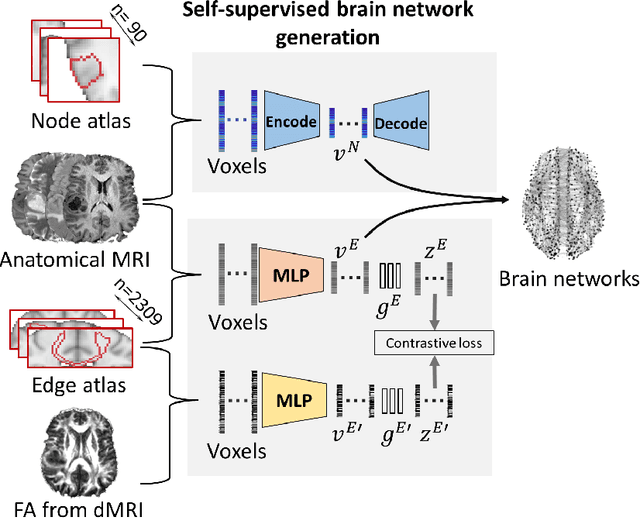

Abstract:The isocitrate dehydrogenase (IDH) gene mutation is an essential biomarker for the diagnosis and prognosis of glioma. It is promising to better predict glioma genotype by integrating focal tumor image and geometric features with brain network features derived from MRI. Convolutions neural networks show reasonable performance in predicting IDH mutation, which, however, cannot learn from non-Euclidean data, e.g., geometric and network data. In this study, we propose a multi-modal learning framework using three separate encoders to extract features of focal tumor image, tumor geometrics and global brain networks. To mitigate the limited availability of diffusion MRI, we develop a self-supervised approach to generate brain networks from anatomical multi-sequence MRI. Moreover, to extract tumor-related features from the brain network, we design a hierarchical attention module for the brain network encoder. Further, we design a bi-level multi-modal contrastive loss to align the multi-modal features and tackle the domain gap at the focal tumor and global brain. Finally, we propose a weighted population graph to integrate the multi-modal features for genotype prediction. Experimental results on the testing set show that the proposed model outperforms the baseline deep learning models. The ablation experiments validate the performance of different components of the framework. The visualized interpretation corresponds to clinical knowledge with further validation. In conclusion, the proposed learning framework provides a novel approach for predicting the genotype of glioma.
Mutual Contrastive Learning to Disentangle Whole Slide Image Representations for Glioma Grading
Mar 08, 2022

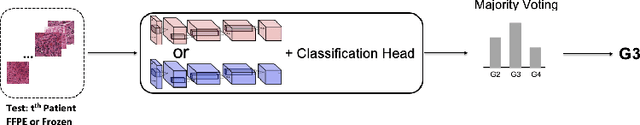
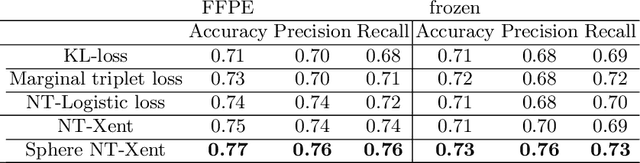
Abstract:Whole slide images (WSI) provide valuable phenotypic information for histological assessment and malignancy grading of tumors. The WSI-based computational pathology promises to provide rapid diagnostic support and facilitate digital health. The most commonly used WSI are derived from formalin-fixed paraffin-embedded (FFPE) and frozen sections. Currently, the majority of automatic tumor grading models are developed based on FFPE sections, which could be affected by the artifacts introduced by tissue processing. Here we propose a mutual contrastive learning scheme to integrate FFPE and frozen sections and disentangle cross-modality representations for glioma grading. We first design a mutual learning scheme to jointly optimize the model training based on FFPE and frozen sections. Further, we develop a multi-modality domain alignment mechanism to ensure semantic consistency in the backbone model training. We finally design a sphere normalized temperature-scaled cross-entropy loss (NT-Xent), which could promote cross-modality representation disentangling of FFPE and frozen sections. Our experiments show that the proposed scheme achieves better performance than the model trained based on each single modality or mixed modalities. The sphere NT-Xent loss outperforms other typical metrics loss functions.
Metastatic Cancer Image Classification Based On Deep Learning Method
Nov 13, 2020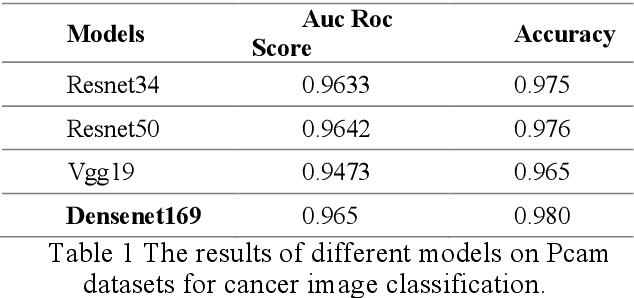
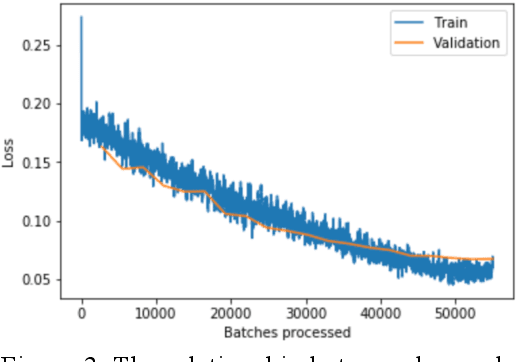
Abstract:Using histopathological images to automatically classify cancer is a difficult task for accurately detecting cancer, especially to identify metastatic cancer in small image patches obtained from larger digital pathology scans. Computer diagnosis technology has attracted wide attention from researchers. In this paper, we propose a noval method which combines the deep learning algorithm in image classification, the DenseNet169 framework and Rectified Adam optimization algorithm. The connectivity pattern of DenseNet is direct connections from any layer to all consecutive layers, which can effectively improve the information flow between different layers. With the fact that RAdam is not easy to fall into a local optimal solution, and it can converge quickly in model training. The experimental results shows that our model achieves superior performance over the other classical convolutional neural networks approaches, such as Vgg19, Resnet34, Resnet50. In particular, the Auc-Roc score of our DenseNet169 model is 1.77% higher than Vgg19 model, and the Accuracy score is 1.50% higher. Moreover, we also study the relationship between loss value and batches processed during the training stage and validation stage, and obtain some important and interesting findings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge