Yiran Wei
Stephanie: Step-by-Step Dialogues for Mimicking Human Interactions in Social Conversations
Jul 04, 2024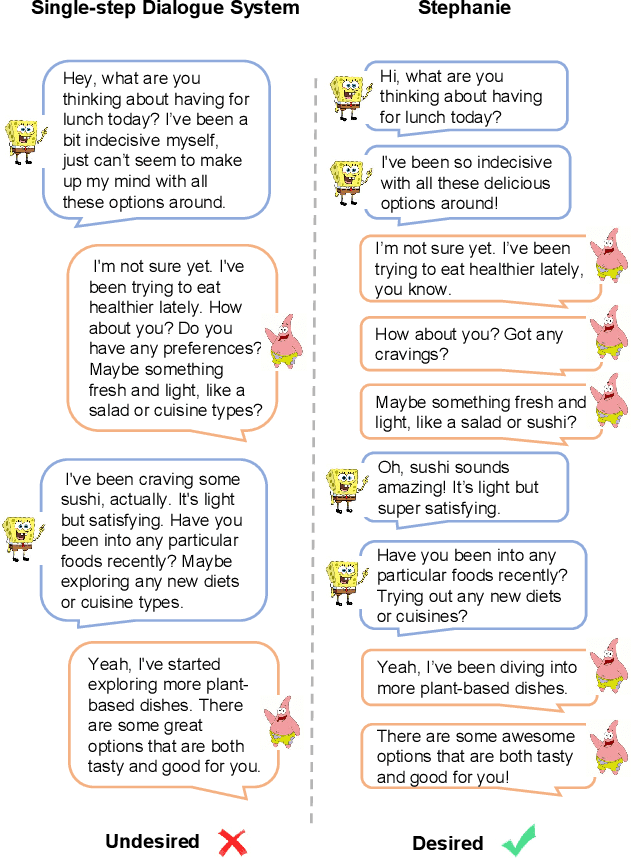
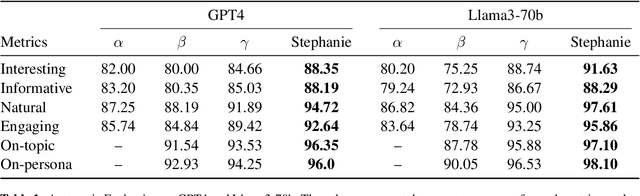
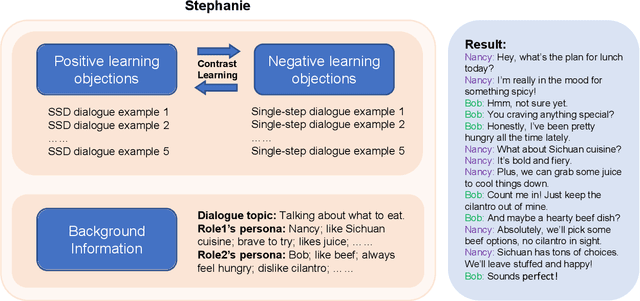
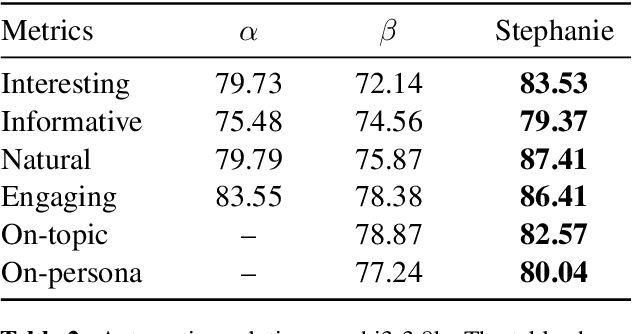
Abstract:In the rapidly evolving field of natural language processing, dialogue systems primarily employ a single-step dialogue paradigm. Although this paradigm is efficient, it lacks the depth and fluidity of human interactions and does not appear natural. We introduce a novel \textbf{Step}-by-Step Dialogue Paradigm (Stephanie), designed to mimic the ongoing dynamic nature of human conversations. By employing a dual learning strategy and a further-split post-editing method, we generated and utilized a high-quality step-by-step dialogue dataset to fine-tune existing large language models, enabling them to perform step-by-step dialogues. We thoroughly present Stephanie. Tailored automatic and human evaluations are conducted to assess its effectiveness compared to the traditional single-step dialogue paradigm. We will release code, Stephanie datasets, and Stephanie LLMs to facilitate the future of chatbot eras.
Letter to the Editor: What are the legal and ethical considerations of submitting radiology reports to ChatGPT?
May 09, 2024Abstract:This letter critically examines the recent article by Infante et al. assessing the utility of large language models (LLMs) like GPT-4, Perplexity, and Bard in identifying urgent findings in emergency radiology reports. While acknowledging the potential of LLMs in generating labels for computer vision, concerns are raised about the ethical implications of using patient data without explicit approval, highlighting the necessity of stringent data protection measures under GDPR.
A self-supervised text-vision framework for automated brain abnormality detection
May 05, 2024



Abstract:Artificial neural networks trained on large, expert-labelled datasets are considered state-of-the-art for a range of medical image recognition tasks. However, categorically labelled datasets are time-consuming to generate and constrain classification to a pre-defined, fixed set of classes. For neuroradiological applications in particular, this represents a barrier to clinical adoption. To address these challenges, we present a self-supervised text-vision framework that learns to detect clinically relevant abnormalities in brain MRI scans by directly leveraging the rich information contained in accompanying free-text neuroradiology reports. Our training approach consisted of two-steps. First, a dedicated neuroradiological language model - NeuroBERT - was trained to generate fixed-dimensional vector representations of neuroradiology reports (N = 50,523) via domain-specific self-supervised learning tasks. Next, convolutional neural networks (one per MRI sequence) learnt to map individual brain scans to their corresponding text vector representations by optimising a mean square error loss. Once trained, our text-vision framework can be used to detect abnormalities in unreported brain MRI examinations by scoring scans against suitable query sentences (e.g., 'there is an acute stroke', 'there is hydrocephalus' etc.), enabling a range of classification-based applications including automated triage. Potentially, our framework could also serve as a clinical decision support tool, not only by suggesting findings to radiologists and detecting errors in provisional reports, but also by retrieving and displaying examples of pathologies from historical examinations that could be relevant to the current case based on textual descriptors.
CLEAN-EVAL: Clean Evaluation on Contaminated Large Language Models
Nov 15, 2023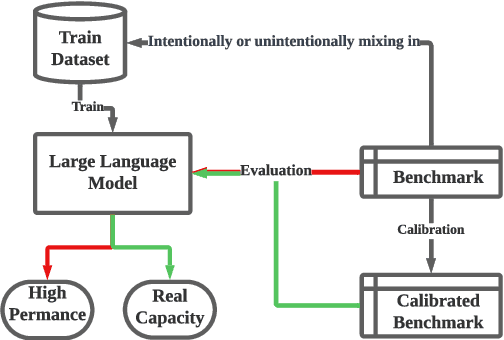
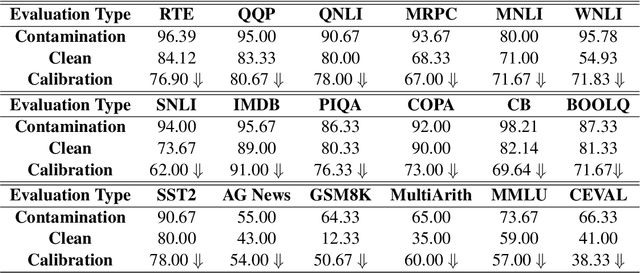
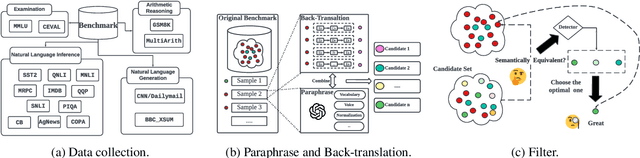
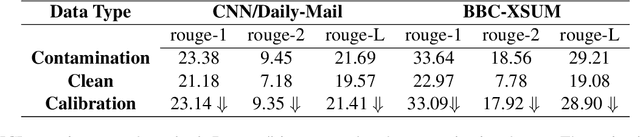
Abstract:We are currently in an era of fierce competition among various large language models (LLMs) continuously pushing the boundaries of benchmark performance. However, genuinely assessing the capabilities of these LLMs has become a challenging and critical issue due to potential data contamination, and it wastes dozens of time and effort for researchers and engineers to download and try those contaminated models. To save our precious time, we propose a novel and useful method, Clean-Eval, which mitigates the issue of data contamination and evaluates the LLMs in a cleaner manner. Clean-Eval employs an LLM to paraphrase and back-translate the contaminated data into a candidate set, generating expressions with the same meaning but in different surface forms. A semantic detector is then used to filter the generated low-quality samples to narrow down this candidate set. The best candidate is finally selected from this set based on the BLEURT score. According to human assessment, this best candidate is semantically similar to the original contamination data but expressed differently. All candidates can form a new benchmark to evaluate the model. Our experiments illustrate that Clean-Eval substantially restores the actual evaluation results on contaminated LLMs under both few-shot learning and fine-tuning scenarios.
Object Topological Character Acquisition by Inductive Learning
Jun 19, 2023



Abstract:Understanding the shape and structure of objects is undoubtedly extremely important for object recognition, but the most common pattern recognition method currently used is machine learning, which often requires a large number of training data. The problem is that this kind of object-oriented learning lacks a priori knowledge. The amount of training data and the complexity of computations are very large, and it is hard to extract explicit knowledge after learning. This is typically called "knowing how without knowing why". We adopted a method of inductive learning, hoping to derive conceptual knowledge of the shape of an object and its formal representation based on a small number of positive examples. It is clear that implementing object recognition is not based on simple physical features such as colors, edges, textures, etc., but on their common geometry, such as topologies, which are stable, persistent, and essential to recognition. In this paper, a formal representation of topological structure based on object's skeleton (RTS) was proposed and the induction process of "seeking common ground" is realized. This research helps promote the method of object recognition from empiricism to rationalism.
Multi-modal learning for predicting the genotype of glioma
Mar 21, 2022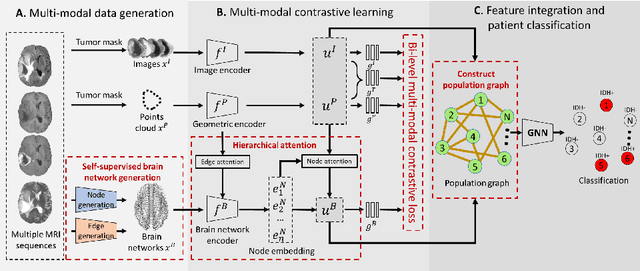
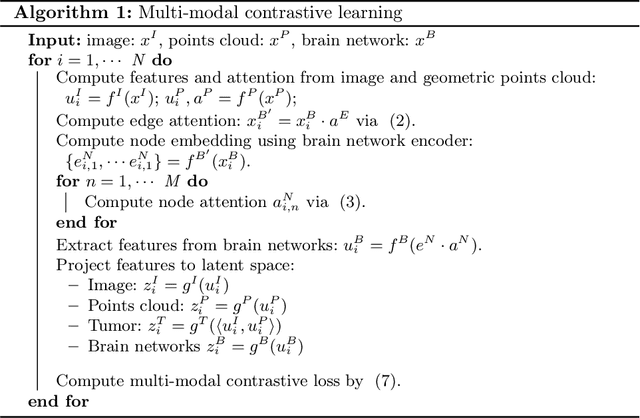
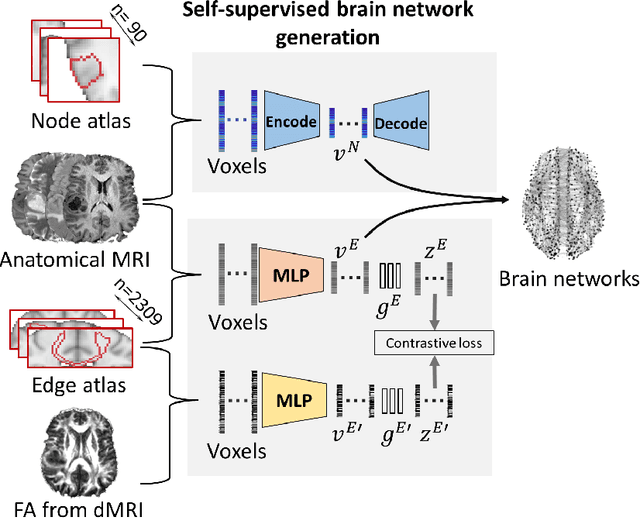

Abstract:The isocitrate dehydrogenase (IDH) gene mutation is an essential biomarker for the diagnosis and prognosis of glioma. It is promising to better predict glioma genotype by integrating focal tumor image and geometric features with brain network features derived from MRI. Convolutions neural networks show reasonable performance in predicting IDH mutation, which, however, cannot learn from non-Euclidean data, e.g., geometric and network data. In this study, we propose a multi-modal learning framework using three separate encoders to extract features of focal tumor image, tumor geometrics and global brain networks. To mitigate the limited availability of diffusion MRI, we develop a self-supervised approach to generate brain networks from anatomical multi-sequence MRI. Moreover, to extract tumor-related features from the brain network, we design a hierarchical attention module for the brain network encoder. Further, we design a bi-level multi-modal contrastive loss to align the multi-modal features and tackle the domain gap at the focal tumor and global brain. Finally, we propose a weighted population graph to integrate the multi-modal features for genotype prediction. Experimental results on the testing set show that the proposed model outperforms the baseline deep learning models. The ablation experiments validate the performance of different components of the framework. The visualized interpretation corresponds to clinical knowledge with further validation. In conclusion, the proposed learning framework provides a novel approach for predicting the genotype of glioma.
Predicting conversion of mild cognitive impairment to Alzheimer's disease
Mar 08, 2022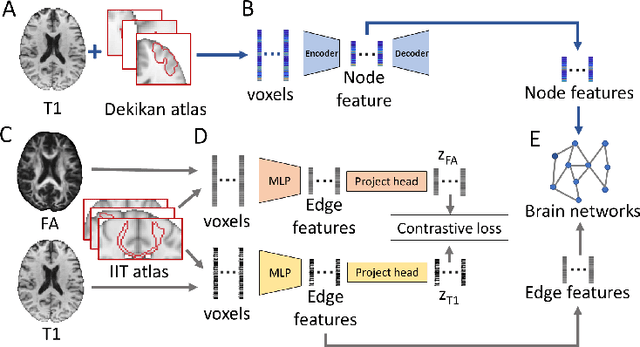
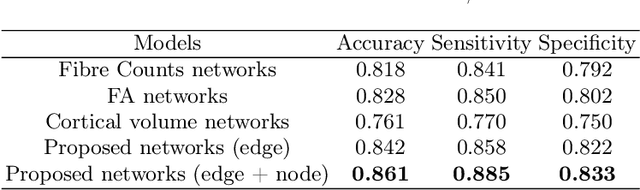
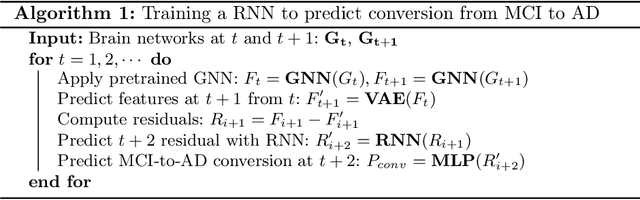
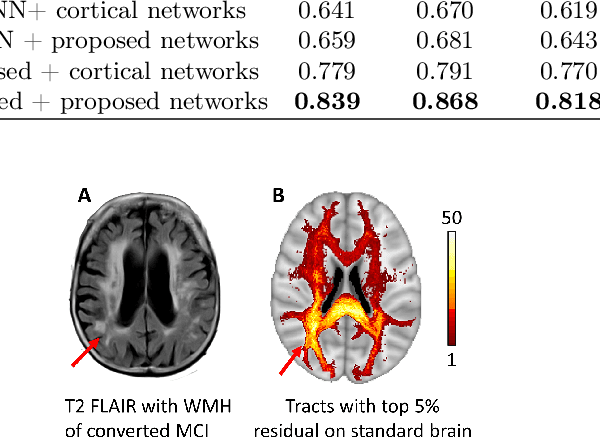
Abstract:Alzheimer's disease (AD) is the most common age-related dementia. Mild cognitive impairment (MCI) is the early stage of cognitive decline before AD. It is crucial to predict the MCI-to-AD conversion for precise management, which remains challenging due to the diversity of patients. Previous evidence shows that the brain network generated from diffusion MRI promises to classify dementia using deep learning. However, the limited availability of diffusion MRI challenges the model training. In this study, we develop a self-supervised contrastive learning approach to generate structural brain networks from routine anatomical MRI under the guidance of diffusion MRI. The generated brain networks are applied to train a learning framework for predicting the MCI-to-AD conversion. Instead of directly modelling the AD brain networks, we train a graph encoder and a variational autoencoder to model the healthy ageing trajectories from brain networks of healthy controls. To predict the MCI-to-AD conversion, we further design a recurrent neural networks based approach to model the longitudinal deviation of patients' brain networks from the healthy ageing trajectory. Numerical results show that the proposed methods outperform the benchmarks in the prediction task. We also visualize the model interpretation to explain the prediction and identify abnormal changes of white matter tracts.
Mutual Contrastive Learning to Disentangle Whole Slide Image Representations for Glioma Grading
Mar 08, 2022

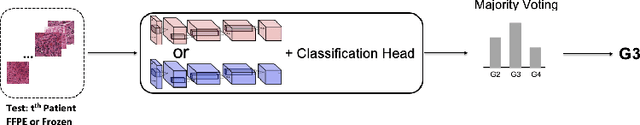
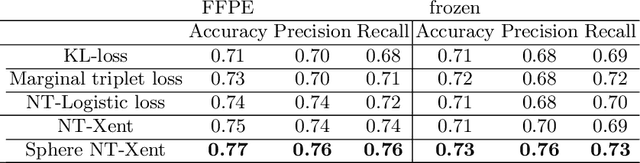
Abstract:Whole slide images (WSI) provide valuable phenotypic information for histological assessment and malignancy grading of tumors. The WSI-based computational pathology promises to provide rapid diagnostic support and facilitate digital health. The most commonly used WSI are derived from formalin-fixed paraffin-embedded (FFPE) and frozen sections. Currently, the majority of automatic tumor grading models are developed based on FFPE sections, which could be affected by the artifacts introduced by tissue processing. Here we propose a mutual contrastive learning scheme to integrate FFPE and frozen sections and disentangle cross-modality representations for glioma grading. We first design a mutual learning scheme to jointly optimize the model training based on FFPE and frozen sections. Further, we develop a multi-modality domain alignment mechanism to ensure semantic consistency in the backbone model training. We finally design a sphere normalized temperature-scaled cross-entropy loss (NT-Xent), which could promote cross-modality representation disentangling of FFPE and frozen sections. Our experiments show that the proposed scheme achieves better performance than the model trained based on each single modality or mixed modalities. The sphere NT-Xent loss outperforms other typical metrics loss functions.
Collaborative learning of images and geometrics for predicting isocitrate dehydrogenase status of glioma
Jan 14, 2022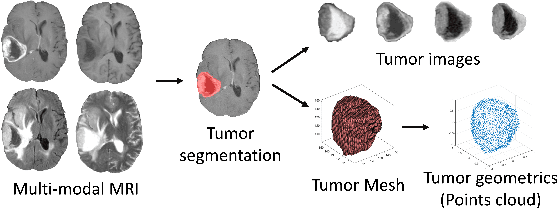

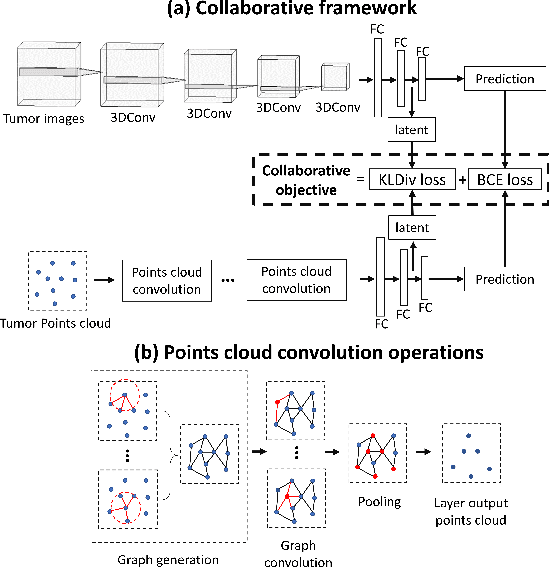
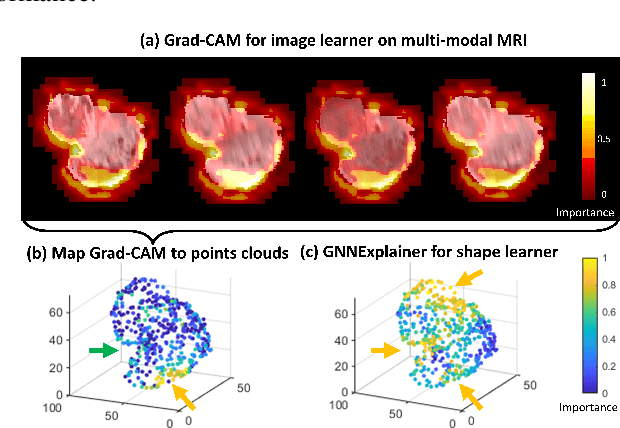
Abstract:The isocitrate dehydrogenase (IDH) gene mutation status is an important biomarker for glioma patients. The gold standard of IDH mutation detection requires tumour tissue obtained via invasive approaches and is usually expensive. Recent advancement in radiogenomics provides a non-invasive approach for predicting IDH mutation based on MRI. Meanwhile, tumor geometrics encompass crucial information for tumour phenotyping. Here we propose a collaborative learning framework that learns both tumor images and tumor geometrics using convolutional neural networks (CNN) and graph neural networks (GNN), respectively. Our results show that the proposed model outperforms the baseline model of 3D-DenseNet121. Further, the collaborative learning model achieves better performance than either the CNN or the GNN alone. The model interpretation shows that the CNN and GNN could identify common and unique regions of interest for IDH mutation prediction. In conclusion, collaborating image and geometric learners provides a novel approach for predicting genotype and characterising glioma.
Predicting isocitrate dehydrogenase mutation status in glioma using structural brain networks and graph neural networks
Sep 10, 2021Abstract:Glioma is a common malignant brain tumor with distinct survival among patients. The isocitrate dehydrogenase (IDH) gene mutation provides critical diagnostic and prognostic value for glioma. It is of crucial significance to non-invasively predict IDH mutation based on pre-treatment MRI. Machine learning/deep learning models show reasonable performance in predicting IDH mutation using MRI. However, most models neglect the systematic brain alterations caused by tumor invasion, where widespread infiltration along white matter tracts is a hallmark of glioma. Structural brain network provides an effective tool to characterize brain organisation, which could be captured by the graph neural networks (GNN) to more accurately predict IDH mutation. Here we propose a method to predict IDH mutation using GNN, based on the structural brain network of patients. Specifically, we firstly construct a network template of healthy subjects, consisting of atlases of edges (white matter tracts) and nodes (cortical/subcortical brain regions) to provide regions of interest (ROIs). Next, we employ autoencoders to extract the latent multi-modal MRI features from the ROIs of edges and nodes in patients, to train a GNN architecture for predicting IDH mutation. The results show that the proposed method outperforms the baseline models using the 3D-CNN and 3D-DenseNet. In addition, model interpretation suggests its ability to identify the tracts infiltrated by tumor, corresponding to clinical prior knowledge. In conclusion, integrating brain networks with GNN offers a new avenue to study brain lesions using computational neuroscience and computer vision approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge