Marc Modat
Machine learning algorithms to predict the risk of rupture of intracranial aneurysms: a systematic review
Dec 06, 2024Abstract:Purpose: Subarachnoid haemorrhage is a potentially fatal consequence of intracranial aneurysm rupture, however, it is difficult to predict if aneurysms will rupture. Prophylactic treatment of an intracranial aneurysm also involves risk, hence identifying rupture-prone aneurysms is of substantial clinical importance. This systematic review aims to evaluate the performance of machine learning algorithms for predicting intracranial aneurysm rupture risk. Methods: MEDLINE, Embase, Cochrane Library and Web of Science were searched until December 2023. Studies incorporating any machine learning algorithm to predict the risk of rupture of an intracranial aneurysm were included. Risk of bias was assessed using the Prediction Model Risk of Bias Assessment Tool (PROBAST). PROSPERO registration: CRD42023452509. Results: Out of 10,307 records screened, 20 studies met the eligibility criteria for this review incorporating a total of 20,286 aneurysm cases. The machine learning models gave a 0.66-0.90 range for performance accuracy. The models were compared to current clinical standards in six studies and gave mixed results. Most studies posed high or unclear risks of bias and concerns for applicability, limiting the inferences that can be drawn from them. There was insufficient homogenous data for a meta-analysis. Conclusions: Machine learning can be applied to predict the risk of rupture for intracranial aneurysms. However, the evidence does not comprehensively demonstrate superiority to existing practice, limiting its role as a clinical adjunct. Further prospective multicentre studies of recent machine learning tools are needed to prove clinical validation before they are implemented in the clinic.
Letter to the Editor: What are the legal and ethical considerations of submitting radiology reports to ChatGPT?
May 09, 2024Abstract:This letter critically examines the recent article by Infante et al. assessing the utility of large language models (LLMs) like GPT-4, Perplexity, and Bard in identifying urgent findings in emergency radiology reports. While acknowledging the potential of LLMs in generating labels for computer vision, concerns are raised about the ethical implications of using patient data without explicit approval, highlighting the necessity of stringent data protection measures under GDPR.
Artificial intelligence for abnormality detection in high volume neuroimaging: a systematic review and meta-analysis
May 09, 2024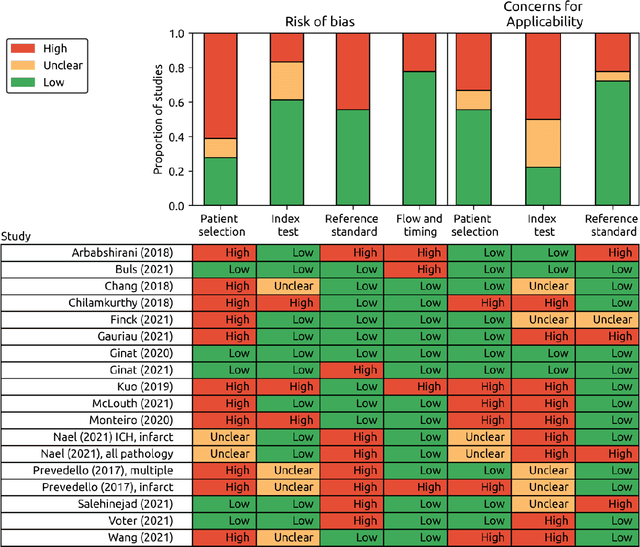
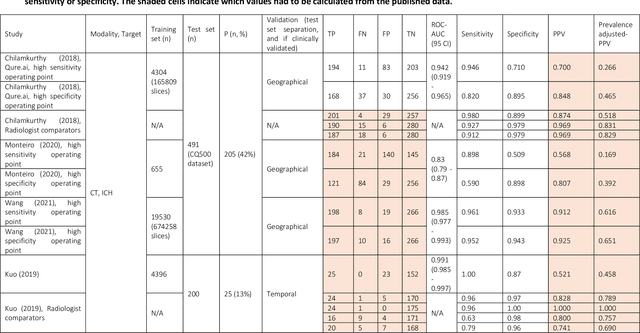
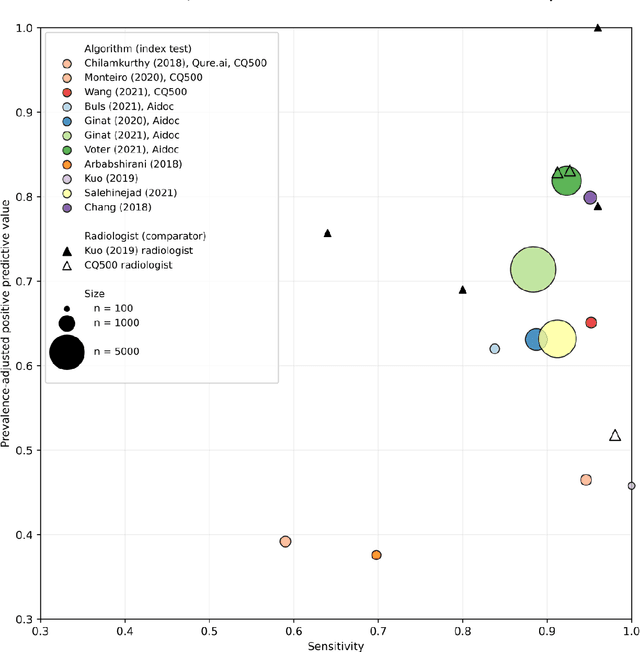
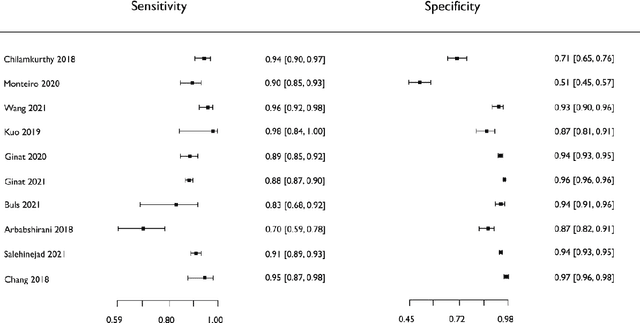
Abstract:Purpose: Most studies evaluating artificial intelligence (AI) models that detect abnormalities in neuroimaging are either tested on unrepresentative patient cohorts or are insufficiently well-validated, leading to poor generalisability to real-world tasks. The aim was to determine the diagnostic test accuracy and summarise the evidence supporting the use of AI models performing first-line, high-volume neuroimaging tasks. Methods: Medline, Embase, Cochrane library and Web of Science were searched until September 2021 for studies that temporally or externally validated AI capable of detecting abnormalities in first-line CT or MR neuroimaging. A bivariate random-effects model was used for meta-analysis where appropriate. PROSPERO: CRD42021269563. Results: Only 16 studies were eligible for inclusion. Included studies were not compromised by unrepresentative datasets or inadequate validation methodology. Direct comparison with radiologists was available in 4/16 studies. 15/16 had a high risk of bias. Meta-analysis was only suitable for intracranial haemorrhage detection in CT imaging (10/16 studies), where AI systems had a pooled sensitivity and specificity 0.90 (95% CI 0.85 - 0.94) and 0.90 (95% CI 0.83 - 0.95) respectively. Other AI studies using CT and MRI detected target conditions other than haemorrhage (2/16), or multiple target conditions (4/16). Only 3/16 studies implemented AI in clinical pathways, either for pre-read triage or as post-read discrepancy identifiers. Conclusion: The paucity of eligible studies reflects that most abnormality detection AI studies were not adequately validated in representative clinical cohorts. The few studies describing how abnormality detection AI could impact patients and clinicians did not explore the full ramifications of clinical implementation.
Generative AI for Medical Imaging: extending the MONAI Framework
Jul 27, 2023Abstract:Recent advances in generative AI have brought incredible breakthroughs in several areas, including medical imaging. These generative models have tremendous potential not only to help safely share medical data via synthetic datasets but also to perform an array of diverse applications, such as anomaly detection, image-to-image translation, denoising, and MRI reconstruction. However, due to the complexity of these models, their implementation and reproducibility can be difficult. This complexity can hinder progress, act as a use barrier, and dissuade the comparison of new methods with existing works. In this study, we present MONAI Generative Models, a freely available open-source platform that allows researchers and developers to easily train, evaluate, and deploy generative models and related applications. Our platform reproduces state-of-art studies in a standardised way involving different architectures (such as diffusion models, autoregressive transformers, and GANs), and provides pre-trained models for the community. We have implemented these models in a generalisable fashion, illustrating that their results can be extended to 2D or 3D scenarios, including medical images with different modalities (like CT, MRI, and X-Ray data) and from different anatomical areas. Finally, we adopt a modular and extensible approach, ensuring long-term maintainability and the extension of current applications for future features.
MONAI: An open-source framework for deep learning in healthcare
Nov 04, 2022



Abstract:Artificial Intelligence (AI) is having a tremendous impact across most areas of science. Applications of AI in healthcare have the potential to improve our ability to detect, diagnose, prognose, and intervene on human disease. For AI models to be used clinically, they need to be made safe, reproducible and robust, and the underlying software framework must be aware of the particularities (e.g. geometry, physiology, physics) of medical data being processed. This work introduces MONAI, a freely available, community-supported, and consortium-led PyTorch-based framework for deep learning in healthcare. MONAI extends PyTorch to support medical data, with a particular focus on imaging, and provide purpose-specific AI model architectures, transformations and utilities that streamline the development and deployment of medical AI models. MONAI follows best practices for software-development, providing an easy-to-use, robust, well-documented, and well-tested software framework. MONAI preserves the simple, additive, and compositional approach of its underlying PyTorch libraries. MONAI is being used by and receiving contributions from research, clinical and industrial teams from around the world, who are pursuing applications spanning nearly every aspect of healthcare.
Driving Points Prediction For Abdominal Probabilistic Registration
Aug 05, 2022



Abstract:Inter-patient abdominal registration has various applications, from pharmakinematic studies to anatomy modeling. Yet, it remains a challenging application due to the morphological heterogeneity and variability of the human abdomen. Among the various registration methods proposed for this task, probabilistic displacement registration models estimate displacement distribution for a subset of points by comparing feature vectors of points from the two images. These probabilistic models are informative and robust while allowing large displacements by design. As the displacement distributions are typically estimated on a subset of points (which we refer to as driving points), due to computational requirements, we propose in this work to learn a driving points predictor. Compared to previously proposed methods, the driving points predictor is optimized in an end-to-end fashion to infer driving points tailored for a specific registration pipeline. We evaluate the impact of our contribution on two different datasets corresponding to different modalities. Specifically, we compared the performances of 6 different probabilistic displacement registration models when using a driving points predictor or one of 2 other standard driving points selection methods. The proposed method improved performances in 11 out of 12 experiments.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
A multi-organ point cloud registration algorithm for abdominal CT registration
Mar 15, 2022



Abstract:Registering CT images of the chest is a crucial step for several tasks such as disease progression tracking or surgical planning. It is also a challenging step because of the heterogeneous content of the human abdomen which implies complex deformations. In this work, we focus on accurately registering a subset of organs of interest. We register organ surface point clouds, as may typically be extracted from an automatic segmentation pipeline, by expanding the Bayesian Coherent Point Drift algorithm (BCPD). We introduce MO-BCPD, a multi-organ version of the BCPD algorithm which explicitly models three important aspects of this task: organ individual elastic properties, inter-organ motion coherence and segmentation inaccuracy. This model also provides an interpolation framework to estimate the deformation of the entire volume. We demonstrate the efficiency of our method by registering different patients from the LITS challenge dataset. The target registration error on anatomical landmarks is almost twice as small for MO-BCPD compared to standard BCPD while imposing the same constraints on individual organs deformation.
Augmentation based unsupervised domain adaptation
Feb 23, 2022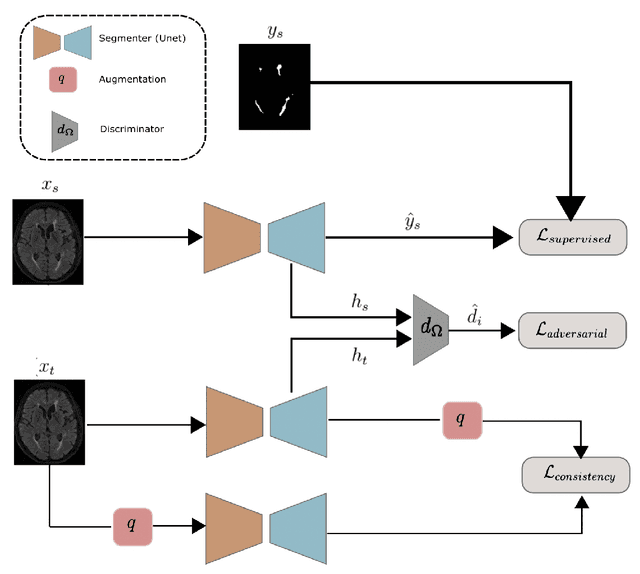
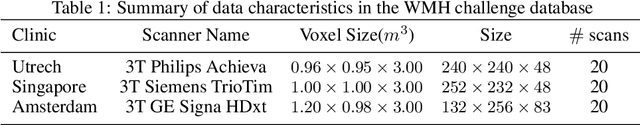
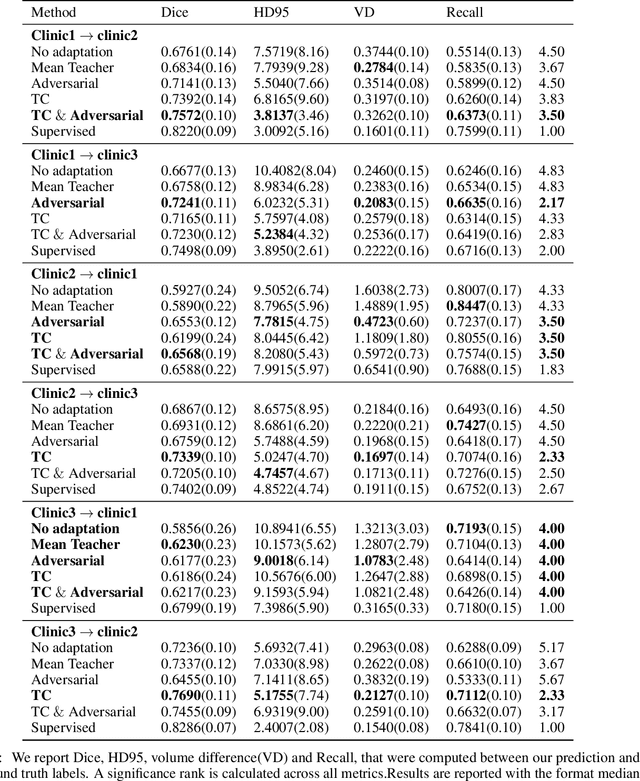
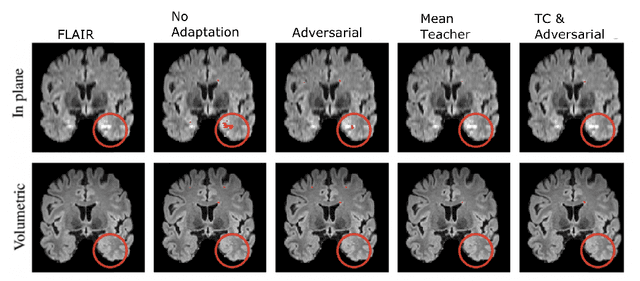
Abstract:The insertion of deep learning in medical image analysis had lead to the development of state-of-the art strategies in several applications such a disease classification, as well as abnormality detection and segmentation. However, even the most advanced methods require a huge and diverse amount of data to generalize. Because in realistic clinical scenarios, data acquisition and annotation is expensive, deep learning models trained on small and unrepresentative data tend to outperform when deployed in data that differs from the one used for training (e.g data from different scanners). In this work, we proposed a domain adaptation methodology to alleviate this problem in segmentation models. Our approach takes advantage of the properties of adversarial domain adaptation and consistency training to achieve more robust adaptation. Using two datasets with white matter hyperintensities (WMH) annotations, we demonstrated that the proposed method improves model generalization even in corner cases where individual strategies tend to fail.
CrossMoDA 2021 challenge: Benchmark of Cross-Modality Domain Adaptation techniques for Vestibular Schwnannoma and Cochlea Segmentation
Jan 08, 2022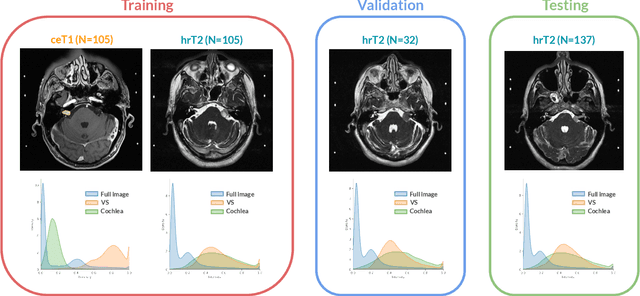
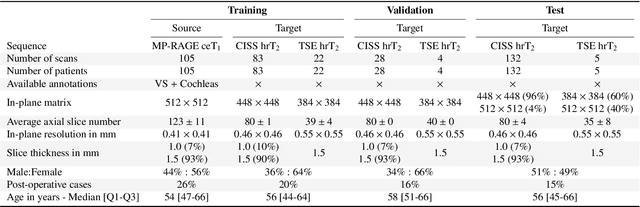
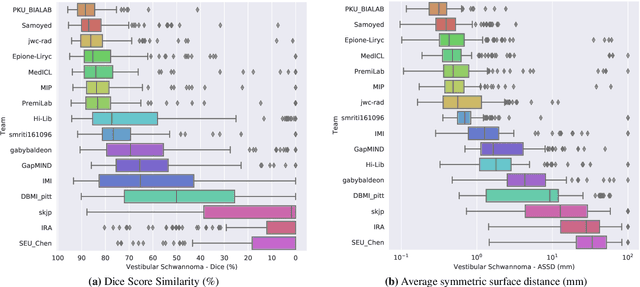
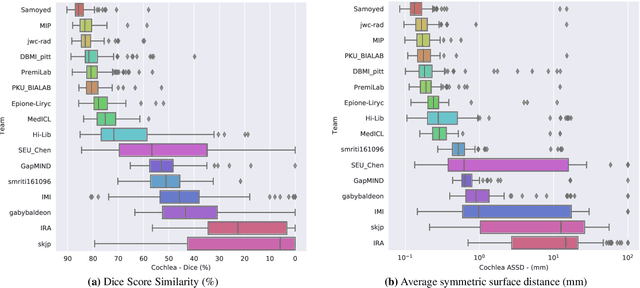
Abstract:Domain Adaptation (DA) has recently raised strong interests in the medical imaging community. While a large variety of DA techniques has been proposed for image segmentation, most of these techniques have been validated either on private datasets or on small publicly available datasets. Moreover, these datasets mostly addressed single-class problems. To tackle these limitations, the Cross-Modality Domain Adaptation (crossMoDA) challenge was organised in conjunction with the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2021). CrossMoDA is the first large and multi-class benchmark for unsupervised cross-modality DA. The challenge's goal is to segment two key brain structures involved in the follow-up and treatment planning of vestibular schwannoma (VS): the VS and the cochleas. Currently, the diagnosis and surveillance in patients with VS are performed using contrast-enhanced T1 (ceT1) MRI. However, there is growing interest in using non-contrast sequences such as high-resolution T2 (hrT2) MRI. Therefore, we created an unsupervised cross-modality segmentation benchmark. The training set provides annotated ceT1 (N=105) and unpaired non-annotated hrT2 (N=105). The aim was to automatically perform unilateral VS and bilateral cochlea segmentation on hrT2 as provided in the testing set (N=137). A total of 16 teams submitted their algorithm for the evaluation phase. The level of performance reached by the top-performing teams is strikingly high (best median Dice - VS:88.4%; Cochleas:85.7%) and close to full supervision (median Dice - VS:92.5%; Cochleas:87.7%). All top-performing methods made use of an image-to-image translation approach to transform the source-domain images into pseudo-target-domain images. A segmentation network was then trained using these generated images and the manual annotations provided for the source image.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge