Akshay Pai
University of Copenhagen, Denmark
Local Gamma Augmentation for Ischemic Stroke Lesion Segmentation on MRI
Jan 12, 2024Abstract:The identification and localisation of pathological tissues in medical images continues to command much attention among deep learning practitioners. When trained on abundant datasets, deep neural networks can match or exceed human performance. However, the scarcity of annotated data complicates the training of these models. Data augmentation techniques can compensate for a lack of training samples. However, many commonly used augmentation methods can fail to provide meaningful samples during model fitting. We present local gamma augmentation, a technique for introducing new instances of intensities in pathological tissues. We leverage local gamma augmentation to compensate for a bias in intensities corresponding to ischemic stroke lesions in human brain MRIs. On three datasets, we show how local gamma augmentation can improve the image-level sensitivity of a deep neural network tasked with ischemic lesion segmentation on magnetic resonance images.
Augmentation based unsupervised domain adaptation
Feb 23, 2022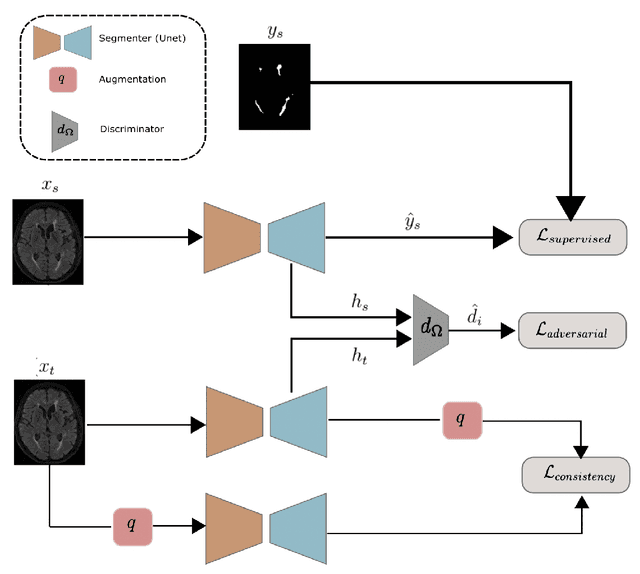
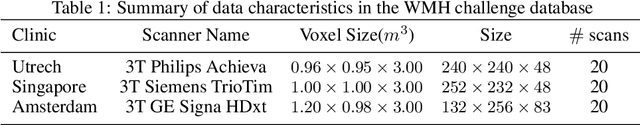
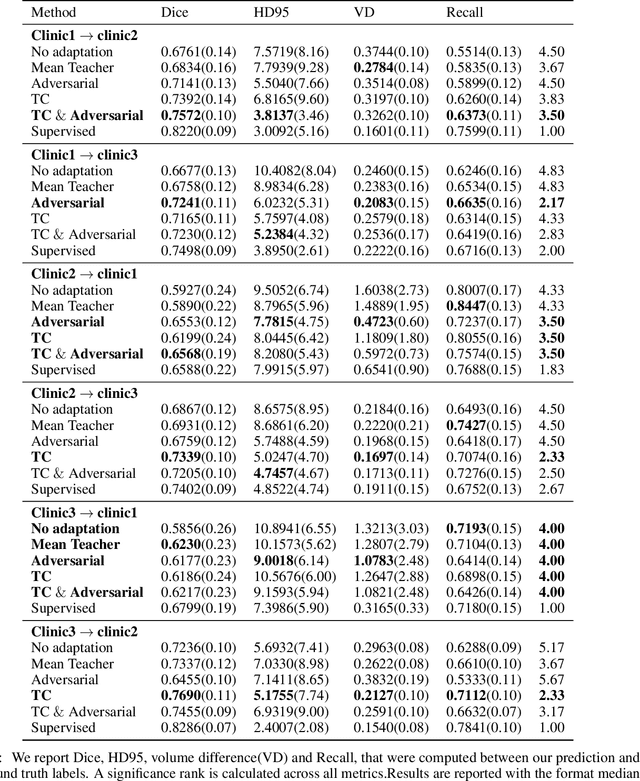
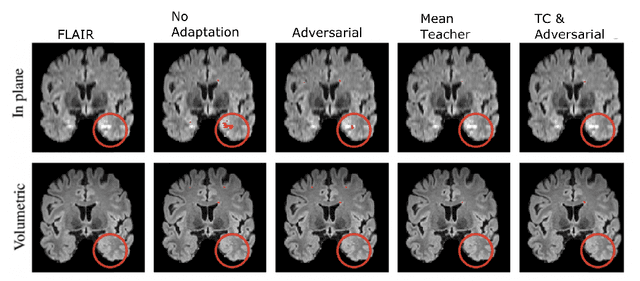
Abstract:The insertion of deep learning in medical image analysis had lead to the development of state-of-the art strategies in several applications such a disease classification, as well as abnormality detection and segmentation. However, even the most advanced methods require a huge and diverse amount of data to generalize. Because in realistic clinical scenarios, data acquisition and annotation is expensive, deep learning models trained on small and unrepresentative data tend to outperform when deployed in data that differs from the one used for training (e.g data from different scanners). In this work, we proposed a domain adaptation methodology to alleviate this problem in segmentation models. Our approach takes advantage of the properties of adversarial domain adaptation and consistency training to achieve more robust adaptation. Using two datasets with white matter hyperintensities (WMH) annotations, we demonstrated that the proposed method improves model generalization even in corner cases where individual strategies tend to fail.
The Medical Segmentation Decathlon
Jun 10, 2021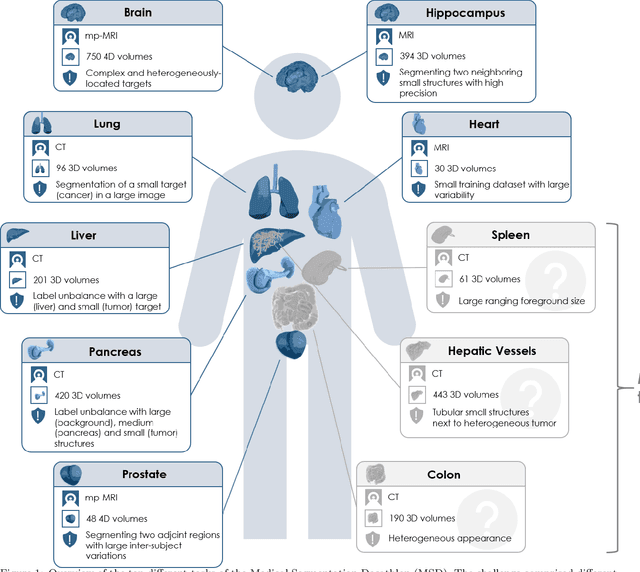
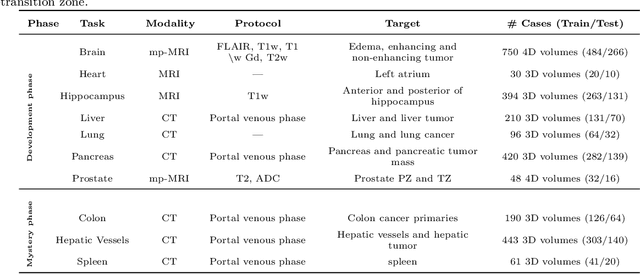
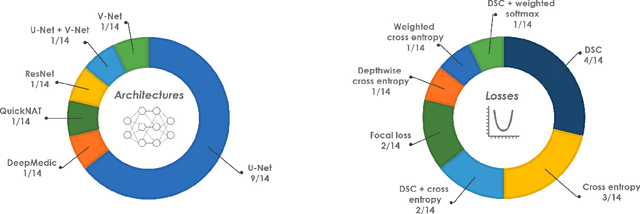
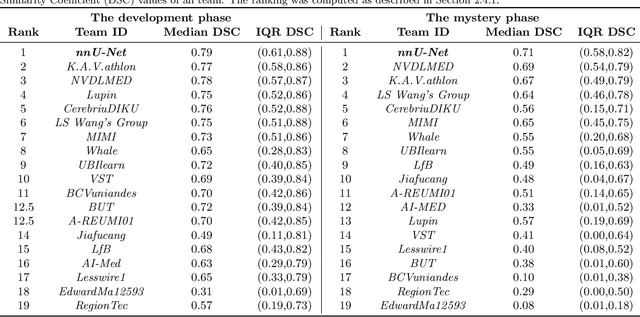
Abstract:International challenges have become the de facto standard for comparative assessment of image analysis algorithms given a specific task. Segmentation is so far the most widely investigated medical image processing task, but the various segmentation challenges have typically been organized in isolation, such that algorithm development was driven by the need to tackle a single specific clinical problem. We hypothesized that a method capable of performing well on multiple tasks will generalize well to a previously unseen task and potentially outperform a custom-designed solution. To investigate the hypothesis, we organized the Medical Segmentation Decathlon (MSD) - a biomedical image analysis challenge, in which algorithms compete in a multitude of both tasks and modalities. The underlying data set was designed to explore the axis of difficulties typically encountered when dealing with medical images, such as small data sets, unbalanced labels, multi-site data and small objects. The MSD challenge confirmed that algorithms with a consistent good performance on a set of tasks preserved their good average performance on a different set of previously unseen tasks. Moreover, by monitoring the MSD winner for two years, we found that this algorithm continued generalizing well to a wide range of other clinical problems, further confirming our hypothesis. Three main conclusions can be drawn from this study: (1) state-of-the-art image segmentation algorithms are mature, accurate, and generalize well when retrained on unseen tasks; (2) consistent algorithmic performance across multiple tasks is a strong surrogate of algorithmic generalizability; (3) the training of accurate AI segmentation models is now commoditized to non AI experts.
Uncertainty quantification in medical image segmentation with Normalizing Flows
Jun 04, 2020



Abstract:Medical image segmentation is inherently an ambiguous task due to factors such as partial volumes and variations in anatomical definitions. While in most cases the segmentation uncertainty is around the border of structures of interest, there can also be considerable inter-rater differences. The class of conditional variational autoencoders (cVAE) offers a principled approach to inferring distributions over plausible segmentations that are conditioned on input images. Segmentation uncertainty estimated from samples of such distributions can be more informative than using pixel level probability scores. In this work, we propose a novel conditional generative model that is based on conditional Normalizing Flow (cFlow). The basic idea is to increase the expressivity of the cVAE by introducing a cFlow transformation step after the encoder. This yields improved approximations of the latent posterior distribution, allowing the model to capture richer segmentation variations. With this we show that the quality and diversity of samples obtained from our conditional generative model is enhanced. Performance of our model, which we call cFlow Net, is evaluated on two medical imaging datasets demonstrating substantial improvements in both qualitative and quantitative measures when compared to a recent cVAE based model.
The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset
May 26, 2020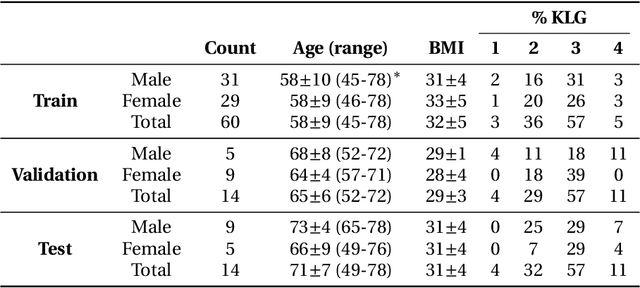
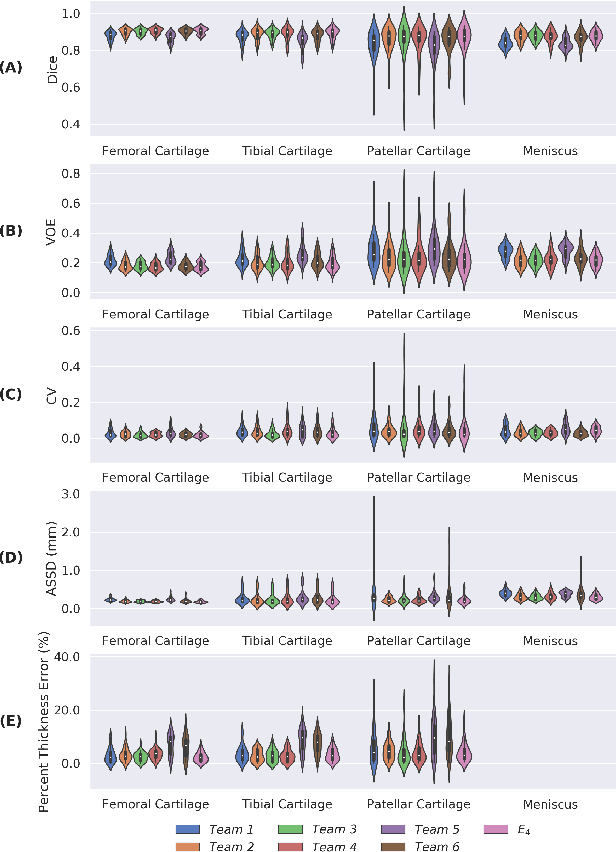
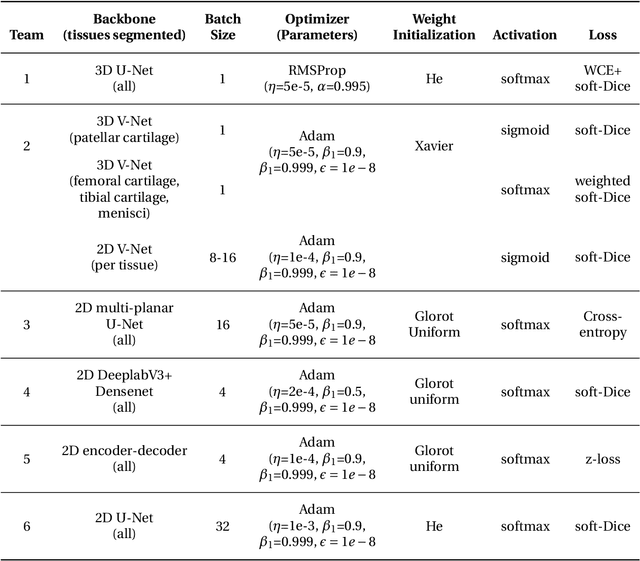
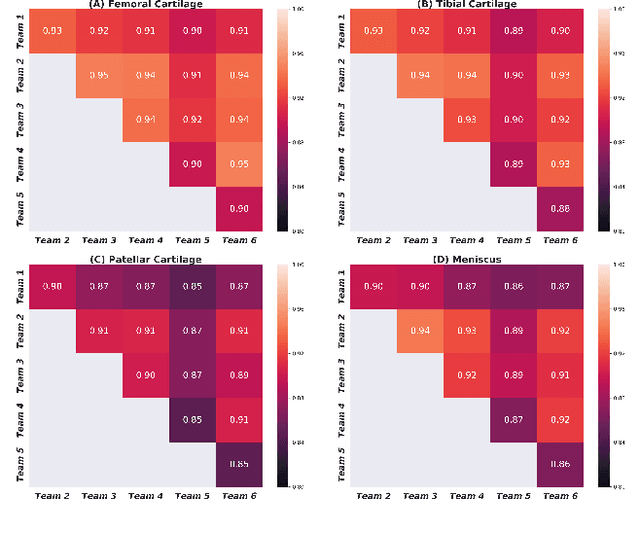
Abstract:Purpose: To organize a knee MRI segmentation challenge for characterizing the semantic and clinical efficacy of automatic segmentation methods relevant for monitoring osteoarthritis progression. Methods: A dataset partition consisting of 3D knee MRI from 88 subjects at two timepoints with ground-truth articular (femoral, tibial, patellar) cartilage and meniscus segmentations was standardized. Challenge submissions and a majority-vote ensemble were evaluated using Dice score, average symmetric surface distance, volumetric overlap error, and coefficient of variation on a hold-out test set. Similarities in network segmentations were evaluated using pairwise Dice correlations. Articular cartilage thickness was computed per-scan and longitudinally. Correlation between thickness error and segmentation metrics was measured using Pearson's coefficient. Two empirical upper bounds for ensemble performance were computed using combinations of model outputs that consolidated true positives and true negatives. Results: Six teams (T1-T6) submitted entries for the challenge. No significant differences were observed across all segmentation metrics for all tissues (p=1.0) among the four top-performing networks (T2, T3, T4, T6). Dice correlations between network pairs were high (>0.85). Per-scan thickness errors were negligible among T1-T4 (p=0.99) and longitudinal changes showed minimal bias (<0.03mm). Low correlations (<0.41) were observed between segmentation metrics and thickness error. The majority-vote ensemble was comparable to top performing networks (p=1.0). Empirical upper bound performances were similar for both combinations (p=1.0). Conclusion: Diverse networks learned to segment the knee similarly where high segmentation accuracy did not correlate to cartilage thickness accuracy. Voting ensembles did not outperform individual networks but may help regularize individual models.
Lung Segmentation from Chest X-rays using Variational Data Imputation
May 20, 2020



Abstract:Pulmonary opacification is the inflammation in the lungs caused by many respiratory ailments, including the novel corona virus disease 2019 (COVID-19). Chest X-rays (CXRs) with such opacifications render regions of lungs imperceptible, making it difficult to perform automated image analysis on them. In this work, we focus on segmenting lungs from such abnormal CXRs as part of a pipeline aimed at automated risk scoring of COVID-19 from CXRs. We treat the high opacity regions as missing data and present a modified CNN-based image segmentation network that utilizes a deep generative model for data imputation. We train this model on normal CXRs with extensive data augmentation and demonstrate the usefulness of this model to extend to cases with extreme abnormalities.
On the Initialization of Long Short-Term Memory Networks
Dec 22, 2019


Abstract:Weight initialization is important for faster convergence and stability of deep neural networks training. In this paper, a robust initialization method is developed to address the training instability in long short-term memory (LSTM) networks. It is based on a normalized random initialization of the network weights that aims at preserving the variance of the network input and output in the same range. The method is applied to standard LSTMs for univariate time series regression and to LSTMs robust to missing values for multivariate disease progression modeling. The results show that in all cases, the proposed initialization method outperforms the state-of-the-art initialization techniques in terms of training convergence and generalization performance of the obtained solution.
One Network to Segment Them All: A General, Lightweight System for Accurate 3D Medical Image Segmentation
Nov 05, 2019


Abstract:Many recent medical segmentation systems rely on powerful deep learning models to solve highly specific tasks. To maximize performance, it is standard practice to evaluate numerous pipelines with varying model topologies, optimization parameters, pre- & postprocessing steps, and even model cascades. It is often not clear how the resulting pipeline transfers to different tasks. We propose a simple and thoroughly evaluated deep learning framework for segmentation of arbitrary medical image volumes. The system requires no task-specific information, no human interaction and is based on a fixed model topology and a fixed hyperparameter set, eliminating the process of model selection and its inherent tendency to cause method-level over-fitting. The system is available in open source and does not require deep learning expertise to use. Without task-specific modifications, the system performed better than or similar to highly specialized deep learning methods across 3 separate segmentation tasks. In addition, it ranked 5-th and 6-th in the first and second round of the 2018 Medical Segmentation Decathlon comprising another 10 tasks. The system relies on multi-planar data augmentation which facilitates the application of a single 2D architecture based on the familiar U-Net. Multi-planar training combines the parameter efficiency of a 2D fully convolutional neural network with a systematic train- and test-time augmentation scheme, which allows the 2D model to learn a representation of the 3D image volume that fosters generalization.
Multi-Domain Adaptation in Brain MRI through Paired Consistency and Adversarial Learning
Sep 17, 2019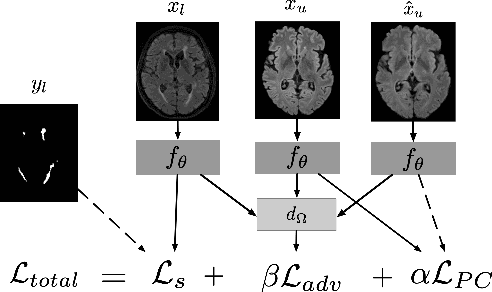
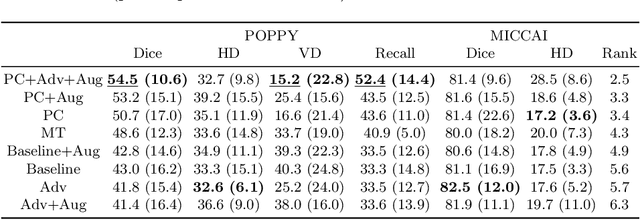
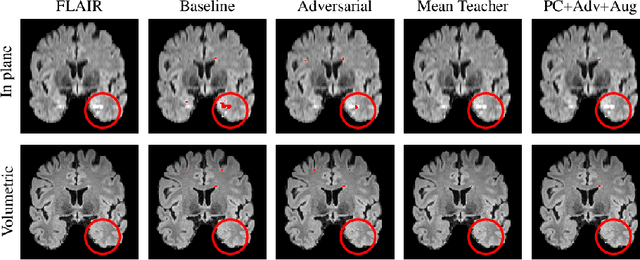
Abstract:Supervised learning algorithms trained on medical images will often fail to generalize across changes in acquisition parameters. Recent work in domain adaptation addresses this challenge and successfully leverages labeled data in a source domain to perform well on an unlabeled target domain. Inspired by recent work in semi-supervised learning we introduce a novel method to adapt from one source domain to $n$ target domains (as long as there is paired data covering all domains). Our multi-domain adaptation method utilises a consistency loss combined with adversarial learning. We provide results on white matter lesion hyperintensity segmentation from brain MRIs using the MICCAI 2017 challenge data as the source domain and two target domains. The proposed method significantly outperforms other domain adaptation baselines.
Knowledge distillation for semi-supervised domain adaptation
Aug 16, 2019
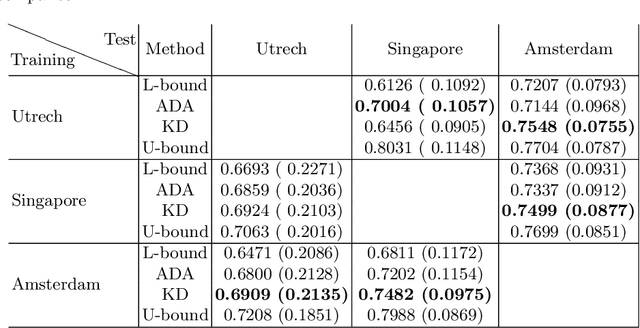
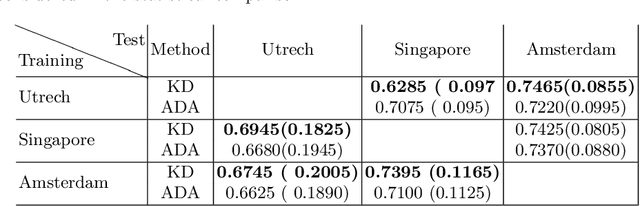
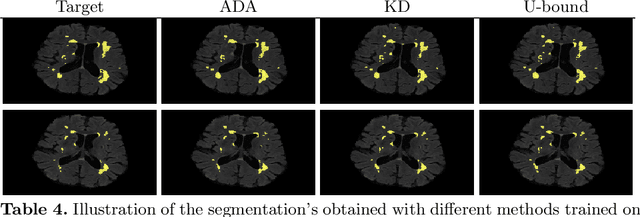
Abstract:In the absence of sufficient data variation (e.g., scanner and protocol variability) in annotated data, deep neural networks (DNNs) tend to overfit during training. As a result, their performance is significantly lower on data from unseen sources compared to the performance on data from the same source as the training data. Semi-supervised domain adaptation methods can alleviate this problem by tuning networks to new target domains without the need for annotated data from these domains. Adversarial domain adaptation (ADA) methods are a popular choice that aim to train networks in such a way that the features generated are domain agnostic. However, these methods require careful dataset-specific selection of hyperparameters such as the complexity of the discriminator in order to achieve a reasonable performance. We propose to use knowledge distillation (KD) -- an efficient way of transferring knowledge between different DNNs -- for semi-supervised domain adaption of DNNs. It does not require dataset-specific hyperparameter tuning, making it generally applicable. The proposed method is compared to ADA for segmentation of white matter hyperintensities (WMH) in magnetic resonance imaging (MRI) scans generated by scanners that are not a part of the training set. Compared with both the baseline DNN (trained on source domain only and without any adaption to target domain) and with using ADA for semi-supervised domain adaptation, the proposed method achieves significantly higher WMH dice scores.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge