Drew Torigian
Predicting Risk of Pulmonary Fibrosis Formation in PASC Patients
May 15, 2025Abstract:While the acute phase of the COVID-19 pandemic has subsided, its long-term effects persist through Post-Acute Sequelae of COVID-19 (PASC), commonly known as Long COVID. There remains substantial uncertainty regarding both its duration and optimal management strategies. PASC manifests as a diverse array of persistent or newly emerging symptoms--ranging from fatigue, dyspnea, and neurologic impairments (e.g., brain fog), to cardiovascular, pulmonary, and musculoskeletal abnormalities--that extend beyond the acute infection phase. This heterogeneous presentation poses substantial challenges for clinical assessment, diagnosis, and treatment planning. In this paper, we focus on imaging findings that may suggest fibrotic damage in the lungs, a critical manifestation characterized by scarring of lung tissue, which can potentially affect long-term respiratory function in patients with PASC. This study introduces a novel multi-center chest CT analysis framework that combines deep learning and radiomics for fibrosis prediction. Our approach leverages convolutional neural networks (CNNs) and interpretable feature extraction, achieving 82.2% accuracy and 85.5% AUC in classification tasks. We demonstrate the effectiveness of Grad-CAM visualization and radiomics-based feature analysis in providing clinically relevant insights for PASC-related lung fibrosis prediction. Our findings highlight the potential of deep learning-driven computational methods for early detection and risk assessment of PASC-related lung fibrosis--presented for the first time in the literature.
Eyes Tell the Truth: GazeVal Highlights Shortcomings of Generative AI in Medical Imaging
Mar 26, 2025



Abstract:The demand for high-quality synthetic data for model training and augmentation has never been greater in medical imaging. However, current evaluations predominantly rely on computational metrics that fail to align with human expert recognition. This leads to synthetic images that may appear realistic numerically but lack clinical authenticity, posing significant challenges in ensuring the reliability and effectiveness of AI-driven medical tools. To address this gap, we introduce GazeVal, a practical framework that synergizes expert eye-tracking data with direct radiological evaluations to assess the quality of synthetic medical images. GazeVal leverages gaze patterns of radiologists as they provide a deeper understanding of how experts perceive and interact with synthetic data in different tasks (i.e., diagnostic or Turing tests). Experiments with sixteen radiologists revealed that 96.6% of the generated images (by the most recent state-of-the-art AI algorithm) were identified as fake, demonstrating the limitations of generative AI in producing clinically accurate images.
XctDiff: Reconstruction of CT Images with Consistent Anatomical Structures from a Single Radiographic Projection Image
Jun 07, 2024



Abstract:In this paper, we present XctDiff, an algorithm framework for reconstructing CT from a single radiograph, which decomposes the reconstruction process into two easily controllable tasks: feature extraction and CT reconstruction. Specifically, we first design a progressive feature extraction strategy that is able to extract robust 3D priors from radiographs. Then, we use the extracted prior information to guide the CT reconstruction in the latent space. Moreover, we design a homogeneous spatial codebook to improve the reconstruction quality further. The experimental results show that our proposed method achieves state-of-the-art reconstruction performance and overcomes the blurring issue. We also apply XctDiff on self-supervised pre-training task. The effectiveness indicates that it has promising additional applications in medical image analysis. The code is available at:https://github.com/qingze-bai/XctDiff
GazeGNN: A Gaze-Guided Graph Neural Network for Disease Classification
May 29, 2023Abstract:The application of eye-tracking techniques in medical image analysis has become increasingly popular in recent years. It collects the visual search patterns of the domain experts, containing much important information about health and disease. Therefore, how to efficiently integrate radiologists' gaze patterns into the diagnostic analysis turns into a critical question. Existing works usually transform gaze information into visual attention maps (VAMs) to supervise the learning process. However, this time-consuming procedure makes it difficult to develop end-to-end algorithms. In this work, we propose a novel gaze-guided graph neural network (GNN), GazeGNN, to perform disease classification from medical scans. In GazeGNN, we create a unified representation graph that models both the image and gaze pattern information. Hence, the eye-gaze information is directly utilized without being converted into VAMs. With this benefit, we develop a real-time, real-world, end-to-end disease classification algorithm for the first time and avoid the noise and time consumption introduced during the VAM preparation. To our best knowledge, GazeGNN is the first work that adopts GNN to integrate image and eye-gaze data. Our experiments on the public chest X-ray dataset show that our proposed method exhibits the best classification performance compared to existing methods.
The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset
May 26, 2020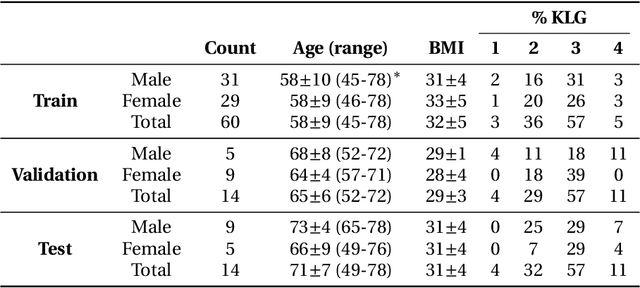
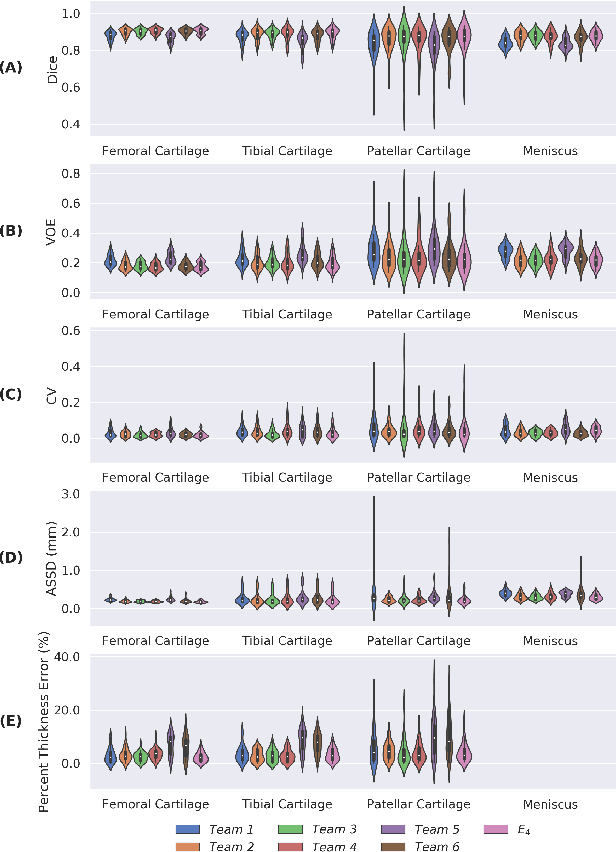
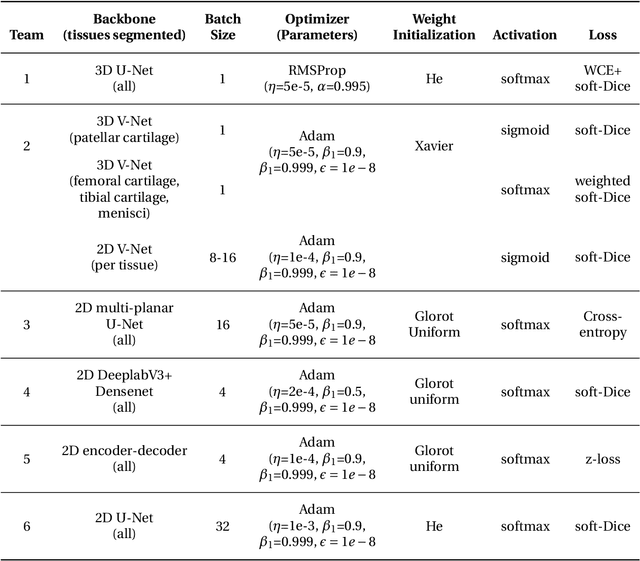
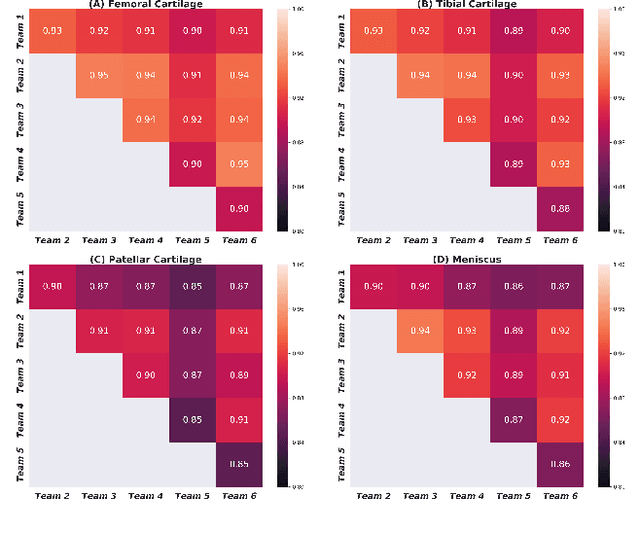
Abstract:Purpose: To organize a knee MRI segmentation challenge for characterizing the semantic and clinical efficacy of automatic segmentation methods relevant for monitoring osteoarthritis progression. Methods: A dataset partition consisting of 3D knee MRI from 88 subjects at two timepoints with ground-truth articular (femoral, tibial, patellar) cartilage and meniscus segmentations was standardized. Challenge submissions and a majority-vote ensemble were evaluated using Dice score, average symmetric surface distance, volumetric overlap error, and coefficient of variation on a hold-out test set. Similarities in network segmentations were evaluated using pairwise Dice correlations. Articular cartilage thickness was computed per-scan and longitudinally. Correlation between thickness error and segmentation metrics was measured using Pearson's coefficient. Two empirical upper bounds for ensemble performance were computed using combinations of model outputs that consolidated true positives and true negatives. Results: Six teams (T1-T6) submitted entries for the challenge. No significant differences were observed across all segmentation metrics for all tissues (p=1.0) among the four top-performing networks (T2, T3, T4, T6). Dice correlations between network pairs were high (>0.85). Per-scan thickness errors were negligible among T1-T4 (p=0.99) and longitudinal changes showed minimal bias (<0.03mm). Low correlations (<0.41) were observed between segmentation metrics and thickness error. The majority-vote ensemble was comparable to top performing networks (p=1.0). Empirical upper bound performances were similar for both combinations (p=1.0). Conclusion: Diverse networks learned to segment the knee similarly where high segmentation accuracy did not correlate to cartilage thickness accuracy. Voting ensembles did not outperform individual networks but may help regularize individual models.
Encoding High-Level Visual Attributes in Capsules for Explainable Medical Diagnoses
Sep 12, 2019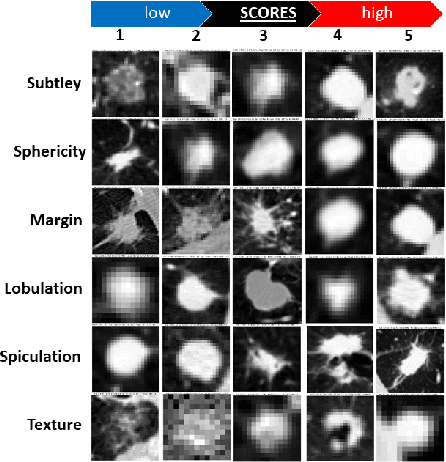
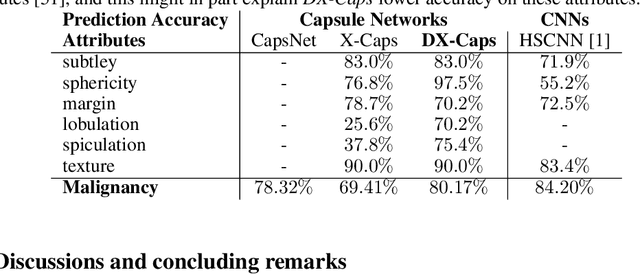
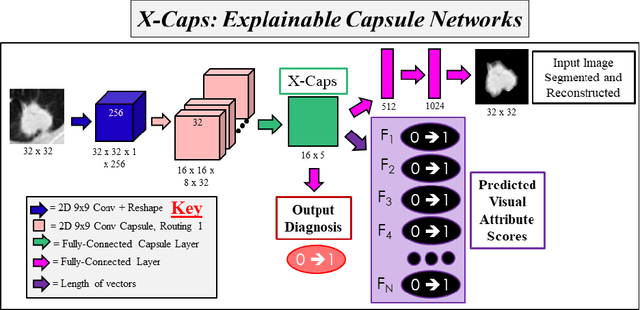
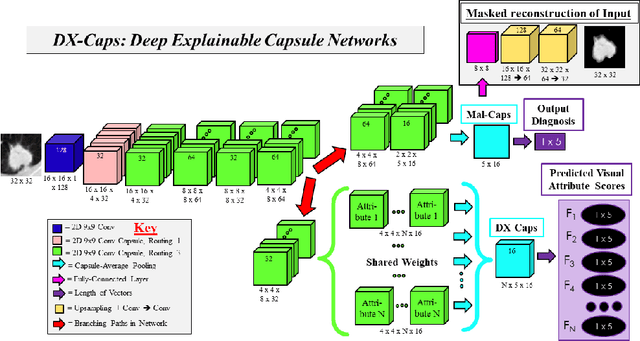
Abstract:Deep neural networks are often called black-boxes due to their difficult-to-interpret decisions. This is characteristic of a deeper trend in machine learning, where predictive performance typically comes at the cost of interpretability. In some domains, such as image-based diagnostic tasks, understanding the reasons behind machine generated predictions is vital in assessing trust. In this study, we introduce novel designs of capsule networks to provide explainable diagnoses. Our proposed deep explainable capsule architecture, called DX-Caps, can encode high-level visual attributes within the vectors of capsules in order to simultaneously produce malignancy predictions for lung cancer as well as approximations of six visually-interpretable attributes, used by radiologists to explain their predictions. To reduce parameter and memory burden of this deeper network, we introduce a new capsule-average pooling function. With this simple, but fundamental addition, capsule networks can be designed in a deeper fashion than was possible before. Our overall approach can be characterized as multi-task learning; we learn to approximate the six high-level visual attributes of a lung nodule within the vectors of our uniquely constructed deep capsule network, while simultaneously segmenting the nodule and predicting its malignancy potential (diagnosis). Tested on over 1000 CT scans, our experimental results show that our proposed algorithm can approximate the visual attributes of lung nodules far better than a deep multi-path dense 3D CNN. The proposed network also achieves higher diagnostic accuracy than a baseline explainable capsule network X-Caps and CapsNet when applied to this task for the first time as well. To the best of our knowledge, this is the first study to investigate capsule networks for visual attribute prediction in general, and explainable medical image diagnosis in particular.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge