Naji Khosravan
Bringing Multi-Modal Multi-Task Federated Foundation Models to Education Domain: Prospects and Challenges
Sep 09, 2025Abstract:Multi-modal multi-task (M3T) foundation models (FMs) have recently shown transformative potential in artificial intelligence, with emerging applications in education. However, their deployment in real-world educational settings is hindered by privacy regulations, data silos, and limited domain-specific data availability. We introduce M3T Federated Foundation Models (FedFMs) for education: a paradigm that integrates federated learning (FL) with M3T FMs to enable collaborative, privacy-preserving training across decentralized institutions while accommodating diverse modalities and tasks. Subsequently, this position paper aims to unveil M3T FedFMs as a promising yet underexplored approach to the education community, explore its potentials, and reveal its related future research directions. We outline how M3T FedFMs can advance three critical pillars of next-generation intelligent education systems: (i) privacy preservation, by keeping sensitive multi-modal student and institutional data local; (ii) personalization, through modular architectures enabling tailored models for students, instructors, and institutions; and (iii) equity and inclusivity, by facilitating participation from underrepresented and resource-constrained entities. We finally identify various open research challenges, including studying of (i) inter-institution heterogeneous privacy regulations, (ii) the non-uniformity of data modalities' characteristics, (iii) the unlearning approaches for M3T FedFMs, (iv) the continual learning frameworks for M3T FedFMs, and (v) M3T FedFM model interpretability, which must be collectively addressed for practical deployment.
Hierarchical Federated Foundation Models over Wireless Networks for Multi-Modal Multi-Task Intelligence: Integration of Edge Learning with D2D/P2P-Enabled Fog Learning Architectures
Sep 03, 2025


Abstract:The rise of foundation models (FMs) has reshaped the landscape of machine learning. As these models continued to grow, leveraging geo-distributed data from wireless devices has become increasingly critical, giving rise to federated foundation models (FFMs). More recently, FMs have evolved into multi-modal multi-task (M3T) FMs (e.g., GPT-4) capable of processing diverse modalities across multiple tasks, which motivates a new underexplored paradigm: M3T FFMs. In this paper, we unveil an unexplored variation of M3T FFMs by proposing hierarchical federated foundation models (HF-FMs), which in turn expose two overlooked heterogeneity dimensions to fog/edge networks that have a direct impact on these emerging models: (i) heterogeneity in collected modalities and (ii) heterogeneity in executed tasks across fog/edge nodes. HF-FMs strategically align the modular structure of M3T FMs, comprising modality encoders, prompts, mixture-of-experts (MoEs), adapters, and task heads, with the hierarchical nature of fog/edge infrastructures. Moreover, HF-FMs enable the optional usage of device-to-device (D2D) communications, enabling horizontal module relaying and localized cooperative training among nodes when feasible. Through delving into the architectural design of HF-FMs, we highlight their unique capabilities along with a series of tailored future research directions. Finally, to demonstrate their potential, we prototype HF-FMs in a wireless network setting and release the open-source code for the development of HF-FMs with the goal of fostering exploration in this untapped field (GitHub: https://github.com/payamsiabd/M3T-FFM).
Multi-Modal Multi-Task (M3T) Federated Foundation Models for Embodied AI: Potentials and Challenges for Edge Integration
May 16, 2025Abstract:As embodied AI systems become increasingly multi-modal, personalized, and interactive, they must learn effectively from diverse sensory inputs, adapt continually to user preferences, and operate safely under resource and privacy constraints. These challenges expose a pressing need for machine learning models capable of swift, context-aware adaptation while balancing model generalization and personalization. Here, two methods emerge as suitable candidates, each offering parts of these capabilities: Foundation Models (FMs) provide a pathway toward generalization across tasks and modalities, whereas Federated Learning (FL) offers the infrastructure for distributed, privacy-preserving model updates and user-level model personalization. However, when used in isolation, each of these approaches falls short of meeting the complex and diverse capability requirements of real-world embodied environments. In this vision paper, we introduce Federated Foundation Models (FFMs) for embodied AI, a new paradigm that unifies the strengths of multi-modal multi-task (M3T) FMs with the privacy-preserving distributed nature of FL, enabling intelligent systems at the wireless edge. We collect critical deployment dimensions of FFMs in embodied AI ecosystems under a unified framework, which we name "EMBODY": Embodiment heterogeneity, Modality richness and imbalance, Bandwidth and compute constraints, On-device continual learning, Distributed control and autonomy, and Yielding safety, privacy, and personalization. For each, we identify concrete challenges and envision actionable research directions. We also present an evaluation framework for deploying FFMs in embodied AI systems, along with the associated trade-offs.
Redefining non-IID Data in Federated Learning for Computer Vision Tasks: Migrating from Labels to Embeddings for Task-Specific Data Distributions
Mar 17, 2025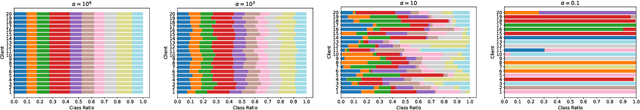
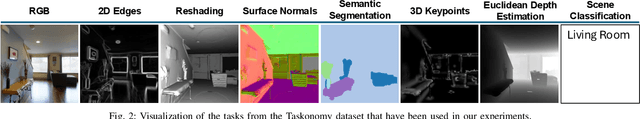
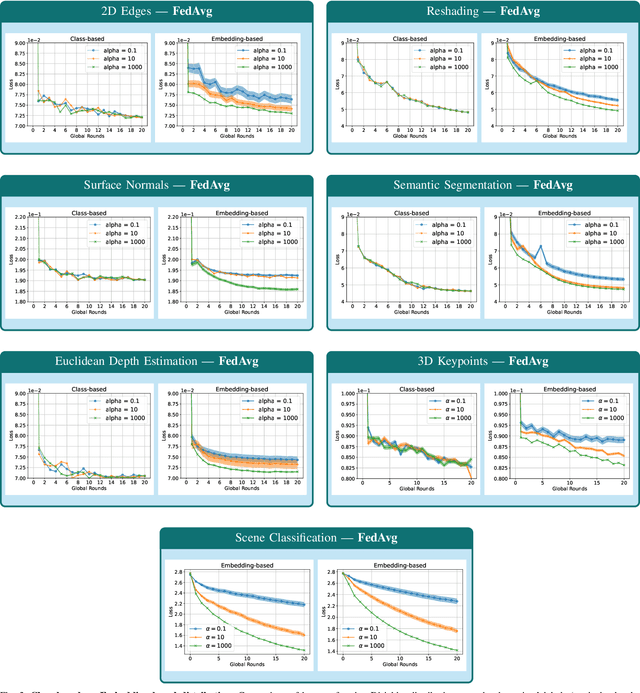
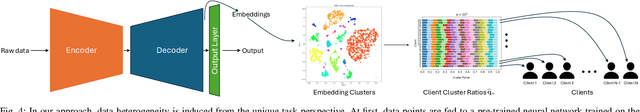
Abstract:Federated Learning (FL) represents a paradigm shift in distributed machine learning (ML), enabling clients to train models collaboratively while keeping their raw data private. This paradigm shift from traditional centralized ML introduces challenges due to the non-iid (non-independent and identically distributed) nature of data across clients, significantly impacting FL's performance. Existing literature, predominantly model data heterogeneity by imposing label distribution skew across clients. In this paper, we show that label distribution skew fails to fully capture the real-world data heterogeneity among clients in computer vision tasks beyond classification. Subsequently, we demonstrate that current approaches overestimate FL's performance by relying on label/class distribution skew, exposing an overlooked gap in the literature. By utilizing pre-trained deep neural networks to extract task-specific data embeddings, we define task-specific data heterogeneity through the lens of each vision task and introduce a new level of data heterogeneity called embedding-based data heterogeneity. Our methodology involves clustering data points based on embeddings and distributing them among clients using the Dirichlet distribution. Through extensive experiments, we evaluate the performance of different FL methods under our revamped notion of data heterogeneity, introducing new benchmark performance measures to the literature. We further unveil a series of open research directions that can be pursued.
Unsupervised Federated Optimization at the Edge: D2D-Enabled Learning without Labels
Apr 15, 2024



Abstract:Federated learning (FL) is a popular solution for distributed machine learning (ML). While FL has traditionally been studied for supervised ML tasks, in many applications, it is impractical to assume availability of labeled data across devices. To this end, we develop Cooperative Federated unsupervised Contrastive Learning ({\tt CF-CL)} to facilitate FL across edge devices with unlabeled datasets. {\tt CF-CL} employs local device cooperation where either explicit (i.e., raw data) or implicit (i.e., embeddings) information is exchanged through device-to-device (D2D) communications to improve local diversity. Specifically, we introduce a \textit{smart information push-pull} methodology for data/embedding exchange tailored to FL settings with either soft or strict data privacy restrictions. Information sharing is conducted through a probabilistic importance sampling technique at receivers leveraging a carefully crafted reserve dataset provided by transmitters. In the implicit case, embedding exchange is further integrated into the local ML training at the devices via a regularization term incorporated into the contrastive loss, augmented with a dynamic contrastive margin to adjust the volume of latent space explored. Numerical evaluations demonstrate that {\tt CF-CL} leads to alignment of latent spaces learned across devices, results in faster and more efficient global model training, and is effective in extreme non-i.i.d. data distribution settings across devices.
Multi-Modal Federated Learning for Cancer Staging over Non-IID Datasets with Unbalanced Modalities
Jan 07, 2024Abstract:The use of machine learning (ML) for cancer staging through medical image analysis has gained substantial interest across medical disciplines. When accompanied by the innovative federated learning (FL) framework, ML techniques can further overcome privacy concerns related to patient data exposure. Given the frequent presence of diverse data modalities within patient records, leveraging FL in a multi-modal learning framework holds considerable promise for cancer staging. However, existing works on multi-modal FL often presume that all data-collecting institutions have access to all data modalities. This oversimplified approach neglects institutions that have access to only a portion of data modalities within the system. In this work, we introduce a novel FL architecture designed to accommodate not only the heterogeneity of data samples, but also the inherent heterogeneity/non-uniformity of data modalities across institutions. We shed light on the challenges associated with varying convergence speeds observed across different data modalities within our FL system. Subsequently, we propose a solution to tackle these challenges by devising a distributed gradient blending and proximity-aware client weighting strategy tailored for multi-modal FL. To show the superiority of our method, we conduct experiments using The Cancer Genome Atlas program (TCGA) datalake considering different cancer types and three modalities of data: mRNA sequences, histopathological image data, and clinical information.
iBARLE: imBalance-Aware Room Layout Estimation
Aug 29, 2023



Abstract:Room layout estimation predicts layouts from a single panorama. It requires datasets with large-scale and diverse room shapes to train the models. However, there are significant imbalances in real-world datasets including the dimensions of layout complexity, camera locations, and variation in scene appearance. These issues considerably influence the model training performance. In this work, we propose the imBalance-Aware Room Layout Estimation (iBARLE) framework to address these issues. iBARLE consists of (1) Appearance Variation Generation (AVG) module, which promotes visual appearance domain generalization, (2) Complex Structure Mix-up (CSMix) module, which enhances generalizability w.r.t. room structure, and (3) a gradient-based layout objective function, which allows more effective accounting for occlusions in complex layouts. All modules are jointly trained and help each other to achieve the best performance. Experiments and ablation studies based on ZInD~\cite{cruz2021zillow} dataset illustrate that iBARLE has state-of-the-art performance compared with other layout estimation baselines.
Graph-CoVis: GNN-based Multi-view Panorama Global Pose Estimation
Apr 26, 2023



Abstract:In this paper, we address the problem of wide-baseline camera pose estimation from a group of 360$^\circ$ panoramas under upright-camera assumption. Recent work has demonstrated the merit of deep-learning for end-to-end direct relative pose regression in 360$^\circ$ panorama pairs [11]. To exploit the benefits of multi-view logic in a learning-based framework, we introduce Graph-CoVis, which non-trivially extends CoVisPose [11] from relative two-view to global multi-view spherical camera pose estimation. Graph-CoVis is a novel Graph Neural Network based architecture that jointly learns the co-visible structure and global motion in an end-to-end and fully-supervised approach. Using the ZInD [4] dataset, which features real homes presenting wide-baselines, occlusion, and limited visual overlap, we show that our model performs competitively to state-of-the-art approaches.
Embedding Alignment for Unsupervised Federated Learning via Smart Data Exchange
Aug 04, 2022


Abstract:Federated learning (FL) has been recognized as one of the most promising solutions for distributed machine learning (ML). In most of the current literature, FL has been studied for supervised ML tasks, in which edge devices collect labeled data. Nevertheless, in many applications, it is impractical to assume existence of labeled data across devices. To this end, we develop a novel methodology, Cooperative Federated unsupervised Contrastive Learning (CF-CL), for FL across edge devices with unlabeled datasets. CF-CL employs local device cooperation where data are exchanged among devices through device-to-device (D2D) communications to avoid local model bias resulting from non-independent and identically distributed (non-i.i.d.) local datasets. CF-CL introduces a push-pull smart data sharing mechanism tailored to unsupervised FL settings, in which, each device pushes a subset of its local datapoints to its neighbors as reserved data points, and pulls a set of datapoints from its neighbors, sampled through a probabilistic importance sampling technique. We demonstrate that CF-CL leads to (i) alignment of unsupervised learned latent spaces across devices, (ii) faster global convergence, allowing for less frequent global model aggregations; and (iii) is effective in extreme non-i.i.d. data settings across the devices.
LASER: LAtent SpacE Rendering for 2D Visual Localization
Apr 01, 2022
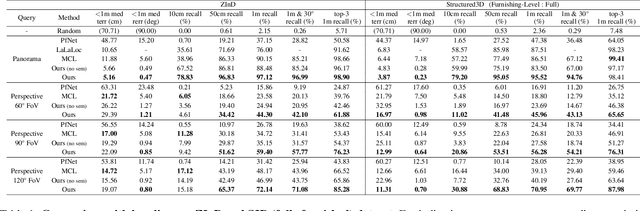

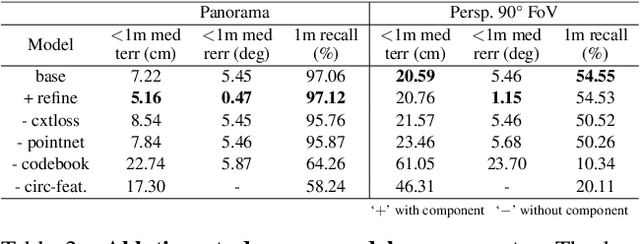
Abstract:We present LASER, an image-based Monte Carlo Localization (MCL) framework for 2D floor maps. LASER introduces the concept of latent space rendering, where 2D pose hypotheses on the floor map are directly rendered into a geometrically-structured latent space by aggregating viewing ray features. Through a tightly coupled rendering codebook scheme, the viewing ray features are dynamically determined at rendering-time based on their geometries (i.e. length, incident-angle), endowing our representation with view-dependent fine-grain variability. Our codebook scheme effectively disentangles feature encoding from rendering, allowing the latent space rendering to run at speeds above 10KHz. Moreover, through metric learning, our geometrically-structured latent space is common to both pose hypotheses and query images with arbitrary field of views. As a result, LASER achieves state-of-the-art performance on large-scale indoor localization datasets (i.e. ZInD and Structured3D) for both panorama and perspective image queries, while significantly outperforming existing learning-based methods in speed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge