Leslie Ying
Knowledge-driven deep learning for fast MR imaging: undersampled MR image reconstruction from supervised to un-supervised learning
Feb 05, 2024Abstract:Deep learning (DL) has emerged as a leading approach in accelerating MR imaging. It employs deep neural networks to extract knowledge from available datasets and then applies the trained networks to reconstruct accurate images from limited measurements. Unlike natural image restoration problems, MR imaging involves physics-based imaging processes, unique data properties, and diverse imaging tasks. This domain knowledge needs to be integrated with data-driven approaches. Our review will introduce the significant challenges faced by such knowledge-driven DL approaches in the context of fast MR imaging along with several notable solutions, which include learning neural networks and addressing different imaging application scenarios. The traits and trends of these techniques have also been given which have shifted from supervised learning to semi-supervised learning, and finally, to unsupervised learning methods. In addition, MR vendors' choices of DL reconstruction have been provided along with some discussions on open questions and future directions, which are critical for the reliable imaging systems.
Multi-Modal Federated Learning for Cancer Staging over Non-IID Datasets with Unbalanced Modalities
Jan 07, 2024Abstract:The use of machine learning (ML) for cancer staging through medical image analysis has gained substantial interest across medical disciplines. When accompanied by the innovative federated learning (FL) framework, ML techniques can further overcome privacy concerns related to patient data exposure. Given the frequent presence of diverse data modalities within patient records, leveraging FL in a multi-modal learning framework holds considerable promise for cancer staging. However, existing works on multi-modal FL often presume that all data-collecting institutions have access to all data modalities. This oversimplified approach neglects institutions that have access to only a portion of data modalities within the system. In this work, we introduce a novel FL architecture designed to accommodate not only the heterogeneity of data samples, but also the inherent heterogeneity/non-uniformity of data modalities across institutions. We shed light on the challenges associated with varying convergence speeds observed across different data modalities within our FL system. Subsequently, we propose a solution to tackle these challenges by devising a distributed gradient blending and proximity-aware client weighting strategy tailored for multi-modal FL. To show the superiority of our method, we conduct experiments using The Cancer Genome Atlas program (TCGA) datalake considering different cancer types and three modalities of data: mRNA sequences, histopathological image data, and clinical information.
Unsupervised Deep Unrolled Reconstruction Using Regularization by Denoising
May 07, 2022



Abstract:Deep learning methods have been successfully used in various computer vision tasks. Inspired by that success, deep learning has been explored in magnetic resonance imaging (MRI) reconstruction. In particular, integrating deep learning and model-based optimization methods has shown considerable advantages. However, a large amount of labeled training data is typically needed for high reconstruction quality, which is challenging for some MRI applications. In this paper, we propose a novel reconstruction method, named DURED-Net, that enables interpretable unsupervised learning for MR image reconstruction by combining an unsupervised denoising network and a plug-and-play method. We aim to boost the reconstruction performance of unsupervised learning by adding an explicit prior that utilizes imaging physics. Specifically, the leverage of a denoising network for MRI reconstruction is achieved using Regularization by Denoising (RED). Experiment results demonstrate that the proposed method requires a reduced amount of training data to achieve high reconstruction quality.
Self-Learned Kernel Low Rank Approach TO Accelerated High Resolution 3D Diffusion MRI
Oct 21, 2021



Abstract:Diffusion Magnetic Resonance Imaging (dMRI) is a promising method to analyze the subtle changes in the tissue structure. However, the lengthy acquisition time is a major limitation in the clinical application of dMRI. Different image acquisition techniques such as parallel imaging, compressed sensing, has shortened the prolonged acquisition time but creating high-resolution 3D dMRI slices still requires a significant amount of time. In this study, we have shown that high-resolution 3D dMRI can be reconstructed from the highly undersampled k-space and q-space data using a Kernel LowRank method. Our proposed method has outperformed the conventional CS methods in terms of both image quality and diffusion maps constructed from the diffusion-weighted images
Deep Manifold Learning for Dynamic MR Imaging
Mar 09, 2021
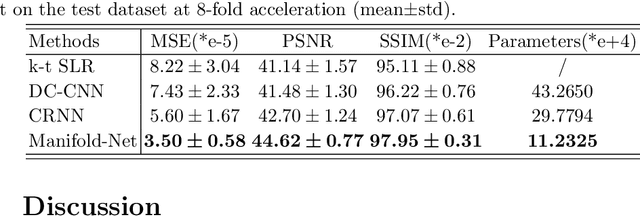
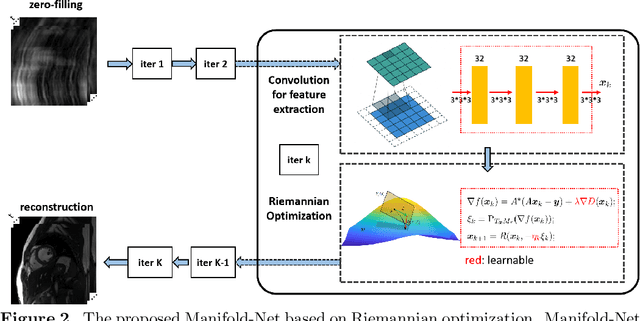

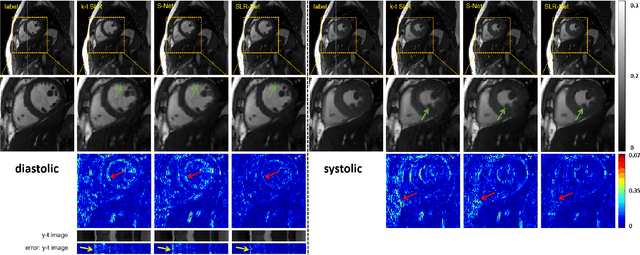
Abstract:Purpose: To develop a deep learning method on a nonlinear manifold to explore the temporal redundancy of dynamic signals to reconstruct cardiac MRI data from highly undersampled measurements. Methods: Cardiac MR image reconstruction is modeled as general compressed sensing (CS) based optimization on a low-rank tensor manifold. The nonlinear manifold is designed to characterize the temporal correlation of dynamic signals. Iterative procedures can be obtained by solving the optimization model on the manifold, including gradient calculation, projection of the gradient to tangent space, and retraction of the tangent space to the manifold. The iterative procedures on the manifold are unrolled to a neural network, dubbed as Manifold-Net. The Manifold-Net is trained using in vivo data with a retrospective electrocardiogram (ECG)-gated segmented bSSFP sequence. Results: Experimental results at high accelerations demonstrate that the proposed method can obtain improved reconstruction compared with a compressed sensing (CS) method k-t SLR and two state-of-the-art deep learning-based methods, DC-CNN and CRNN. Conclusion: This work represents the first study unrolling the optimization on manifolds into neural networks. Specifically, the designed low-rank manifold provides a new technical route for applying low-rank priors in dynamic MR imaging.
Deep Low-rank Prior in Dynamic MR Imaging
Jul 06, 2020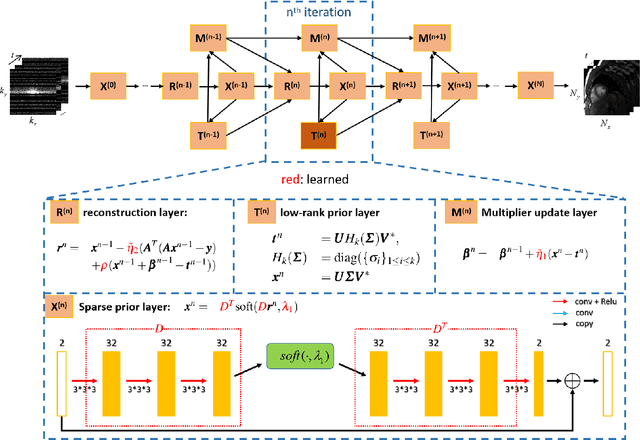
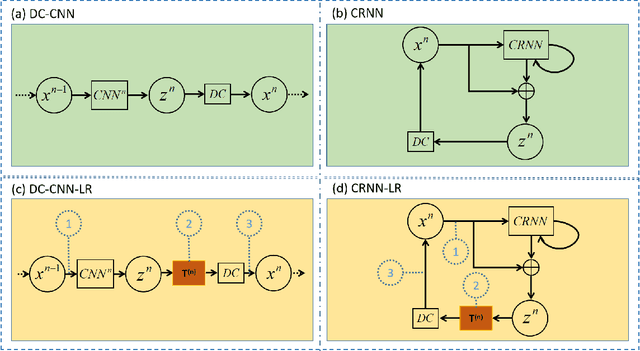
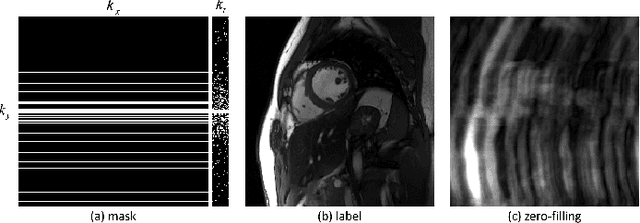
Abstract:The deep learning methods have achieved attractive results in dynamic MR imaging. However, all of these methods only utilize the sparse prior of MR images, while the important low-rank (LR) prior of dynamic MR images is not explored, which limits further improvements of dynamic MR reconstruction. In this paper, a learned singular value thresholding (Learned-SVT) operation is proposed to explore deep low-rank prior in dynamic MR imaging to obtain improved reconstruction results. In particular, we propose two novel and distinct schemes to introduce the learnable low-rank prior into deep network architectures in an unrolling manner and a plug-and-play manner respectively. In the unrolling manner, we propose a model-based unrolling sparse and low-rank network for dynamic MR imaging, dubbed SLR-Net. The SLR-Net is defined over a deep network flow graphs, which is unrolled from the iterative procedures in Iterative Shrinkage-Thresholding Algorithm (ISTA) for optimizing a sparse and low-rank based dynamic MRI model. In the plug-and-play manner, we propose a plug-and-play LR network module that can be easily embedded into any other dynamic MR neural networks without changing the network paradigm. To the best of our knowlegde, this is the first time that a deep low-rank prior has been applied in dynamic MR imaging. Experimental results show that both of the two schemes can further improve the reconstruction results, no matter qualitatively and quantitatively.
Kernel Bi-Linear Modeling for Reconstructing Data on Manifolds: The Dynamic-MRI Case
Feb 27, 2020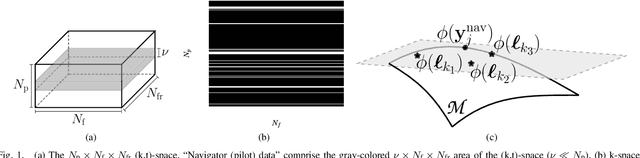
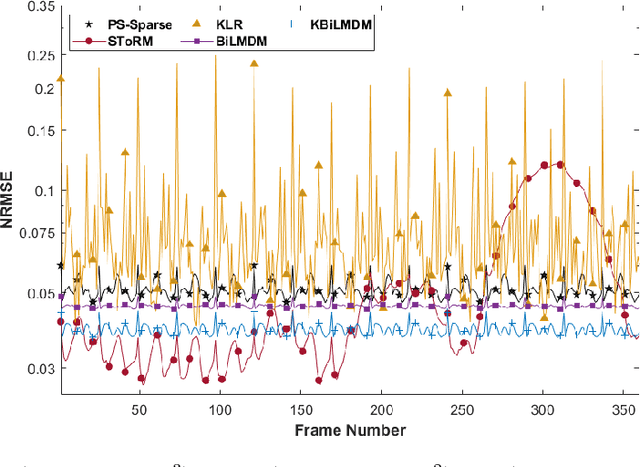
Abstract:This paper establishes a kernel-based framework for reconstructing data on manifolds, tailored to fit the dynamic-(d)MRI-data recovery problem. The proposed methodology exploits simple tangent-space geometries of manifolds in reproducing kernel Hilbert spaces and follows classical kernel-approximation arguments to form the data-recovery task as a bi-linear inverse problem. Departing from mainstream approaches, the proposed methodology uses no training data, employs no graph Laplacian matrix to penalize the optimization task, uses no costly (kernel) pre-imaging step to map feature points back to the input space, and utilizes complex-valued kernel functions to account for k-space data. The framework is validated on synthetically generated dMRI data, where comparisons against state-of-the-art schemes highlight the rich potential of the proposed approach in data-recovery problems.
Deep Learning for Highly Accelerated Diffusion Tensor Imaging
Feb 03, 2020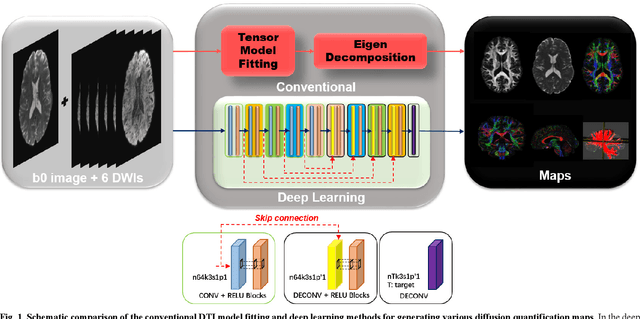
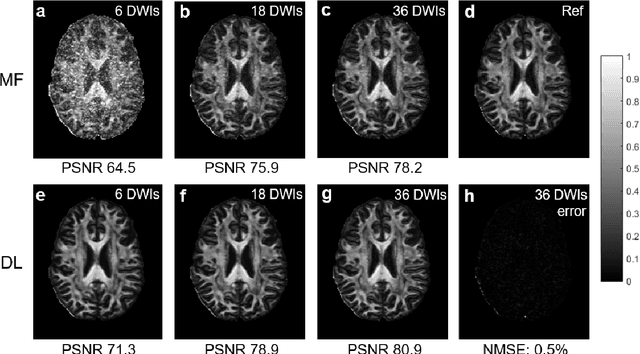
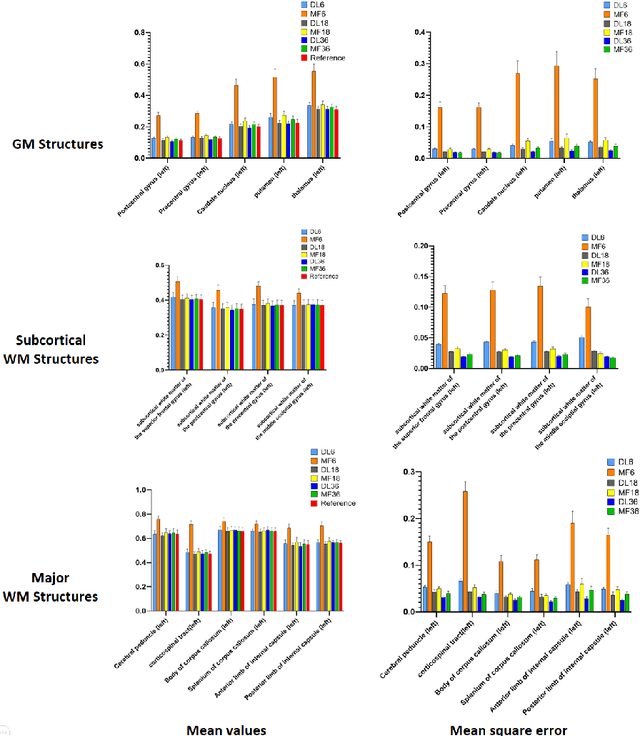

Abstract:Diffusion tensor imaging (DTI) is widely used to examine the human brain white matter structures, including their microarchitecture integrity and spatial fiber tract trajectories, with clinical applications in several neurological disorders and neurosurgical guidance. However, a major factor that prevents DTI from being incorporated in clinical routines is its long scan time due to the acquisition of a large number (typically 30 or more) of diffusion-weighted images (DWIs) required for reliable tensor estimation. Here, a deep learning-based technique is developed to obtain diffusion tensor images with only six DWIs, resulting in a significant reduction in imaging time. The method uses deep convolutional neural networks to learn the highly nonlinear relationship between DWIs and several tensor-derived maps, bypassing the conventional tensor fitting procedure, which is well known to be highly susceptible to noises in DWIs. The performance of the method was evaluated using DWI datasets from the Human Connectome Project and patients with ischemic stroke. Our results demonstrate that the proposed technique is able to generate quantitative maps of good quality fractional anisotropy (FA) and mean diffusivity (MD), as well as the fiber tractography from as few as six DWIs. The proposed method achieves a quantification error of less than 5% in all regions of interest of the brain, which is the rate of in vivo reproducibility of diffusion tensor imaging. Tractography reconstruction is also comparable to the ground truth obtained from 90 DWIs. In addition, we also demonstrate that the neural network trained on healthy volunteers can be directly applied/tested on stroke patients' DWIs data without compromising the lesion detectability. Such a significant reduction in scan time will allow inclusion of DTI into clinical routine for many potential applications.
An Unsupervised Deep Learning Method for Parallel Cardiac MRI via Time-Interleaved Sampling
Dec 20, 2019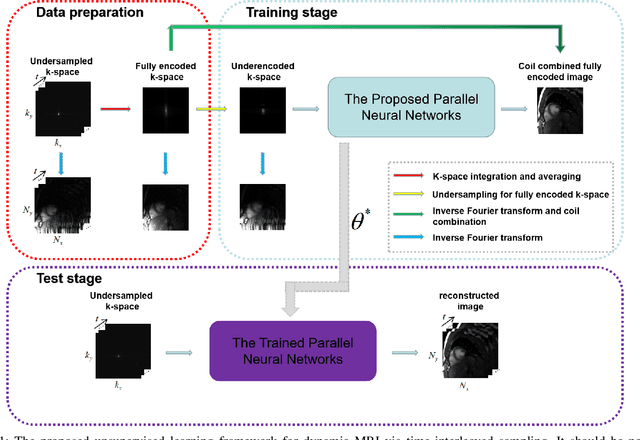
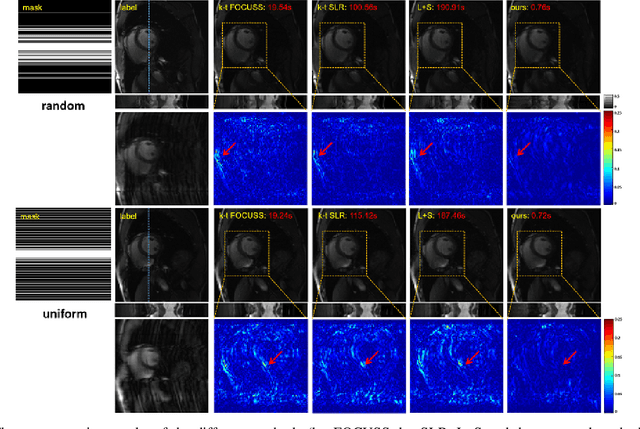
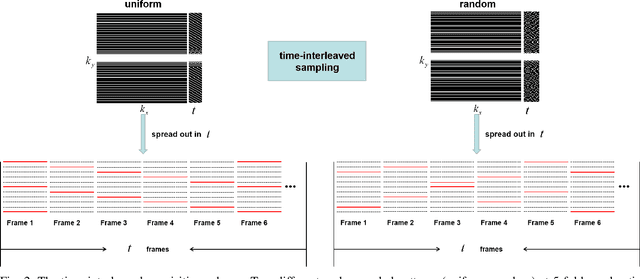
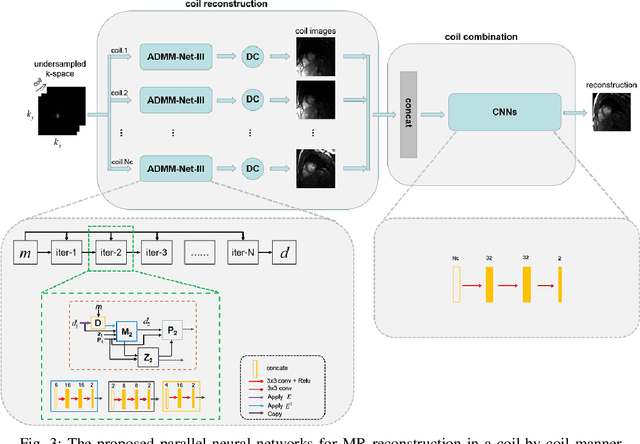
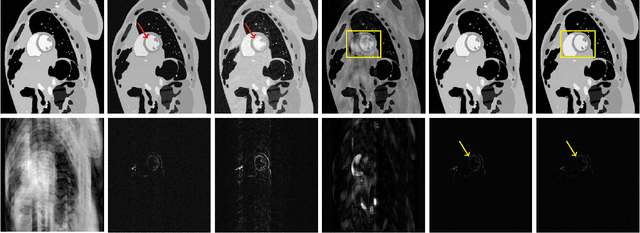
Abstract:Deep learning has achieved good success in cardiac magnetic resonance imaging (MRI) reconstruction, in which convolutional neural networks (CNNs) learn the mapping from undersampled k-space to fully sampled images. Although these deep learning methods can improve reconstruction quality without complex parameter selection or a lengthy reconstruction time compared with iterative methods, the following issues still need to be addressed: 1) all of these methods are based on big data and require a large amount of fully sampled MRI data, which is always difficult for cardiac MRI; 2) All of these methods are only applicable for single-channel images without exploring coil correlation. In this paper, we propose an unsupervised deep learning method for parallel cardiac MRI via a time-interleaved sampling strategy. Specifically, a time-interleaved acquisition scheme is developed to build a set of fully encoded reference data by directly merging the k-space data of adjacent time frames. Then these fully encoded data can be used to train a parallel network for reconstructing images of each coil separately. Finally, the images from each coil are combined together via a CNN to implicitly explore the correlations between coils. The comparisons with classic k-t FOCUSS, k-t SLR and L+S methods on in vivo datasets show that our method can achieve improved reconstruction results in an extremely short amount of time.
Model Learning: Primal Dual Networks for Fast MR imaging
Aug 07, 2019


Abstract:Magnetic resonance imaging (MRI) is known to be a slow imaging modality and undersampling in k-space has been used to increase the imaging speed. However, image reconstruction from undersampled k-space data is an ill-posed inverse problem. Iterative algorithms based on compressed sensing have been used to address the issue. In this work, we unroll the iterations of the primal-dual hybrid gradient algorithm to a learnable deep network architecture, and gradually relax the constraints to reconstruct MR images from highly undersampled k-space data. The proposed method combines the theoretical convergence guarantee of optimi-zation methods with the powerful learning capability of deep networks. As the constraints are gradually relaxed, the reconstruction model is finally learned from the training data by updating in k-space and image domain alternatively. Experi-ments on in vivo MR data demonstrate that the proposed method achieves supe-rior MR reconstructions from highly undersampled k-space data over other state-of-the-art image reconstruction methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge