Ziwen Ke
Zero-shot Dynamic MRI Reconstruction with Global-to-local Diffusion Model
Nov 06, 2024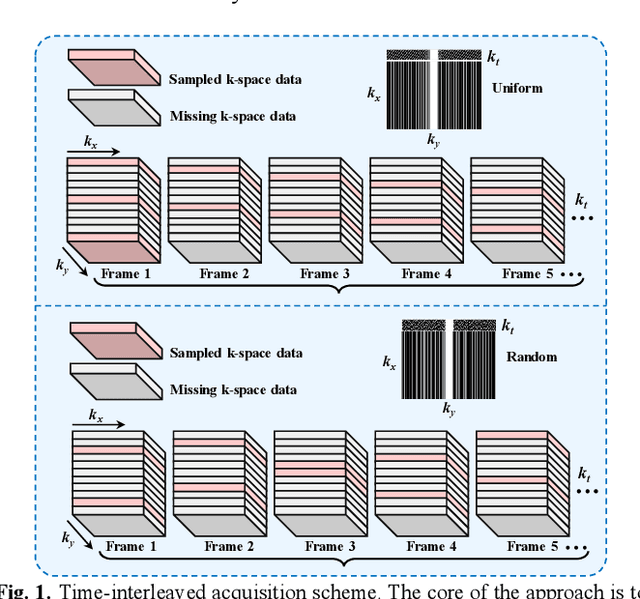
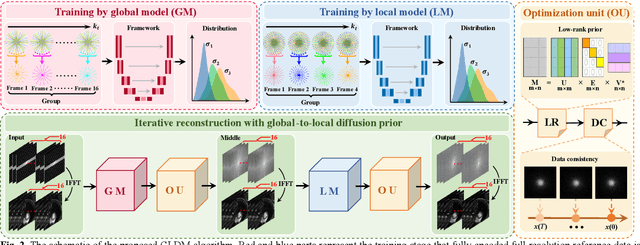
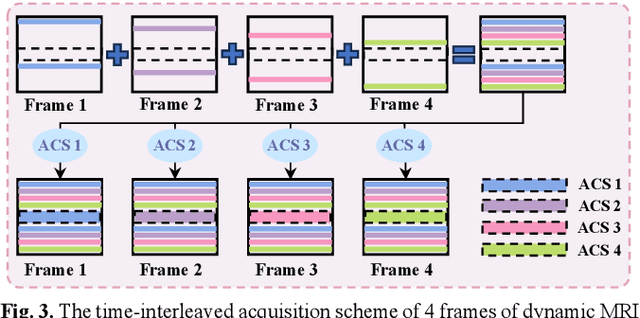
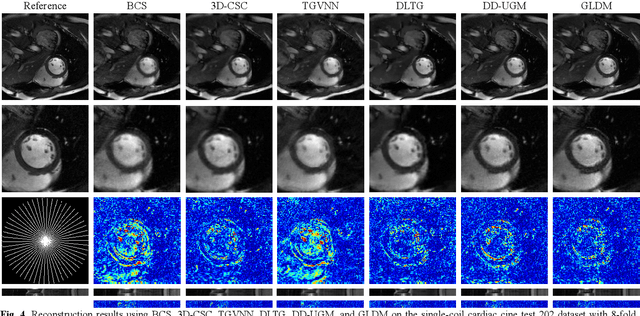
Abstract:Diffusion models have recently demonstrated considerable advancement in the generation and reconstruction of magnetic resonance imaging (MRI) data. These models exhibit great potential in handling unsampled data and reducing noise, highlighting their promise as generative models. However, their application in dynamic MRI remains relatively underexplored. This is primarily due to the substantial amount of fully-sampled data typically required for training, which is difficult to obtain in dynamic MRI due to its spatio-temporal complexity and high acquisition costs. To address this challenge, we propose a dynamic MRI reconstruction method based on a time-interleaved acquisition scheme, termed the Glob-al-to-local Diffusion Model. Specifically, fully encoded full-resolution reference data are constructed by merging under-sampled k-space data from adjacent time frames, generating two distinct bulk training datasets for global and local models. The global-to-local diffusion framework alternately optimizes global information and local image details, enabling zero-shot reconstruction. Extensive experiments demonstrate that the proposed method performs well in terms of noise reduction and detail preservation, achieving reconstruction quality comparable to that of supervised approaches.
Universal Generative Modeling in Dual-domain for Dynamic MR Imaging
Dec 15, 2022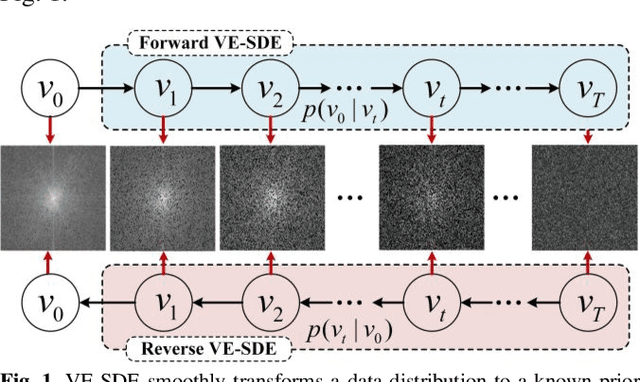
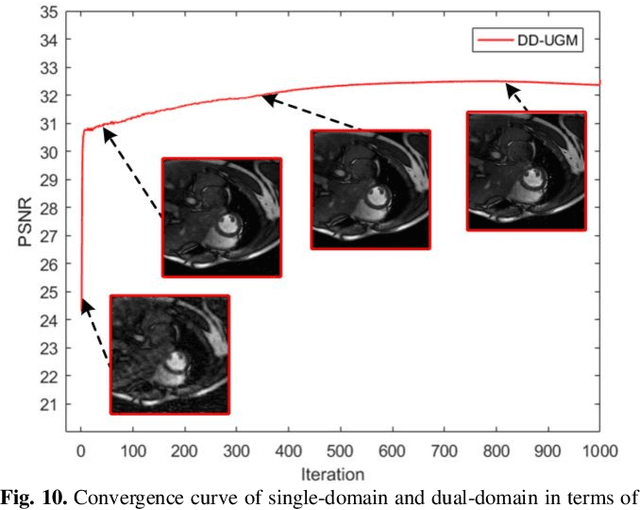
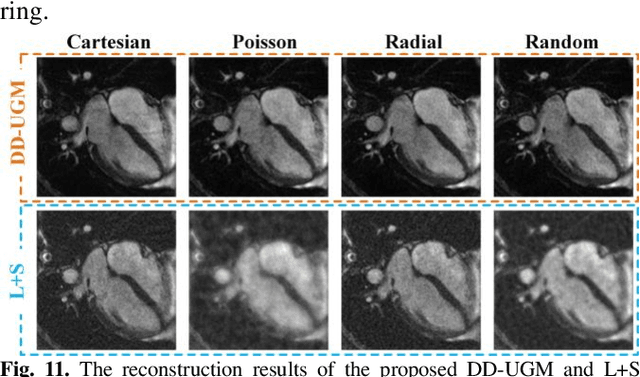
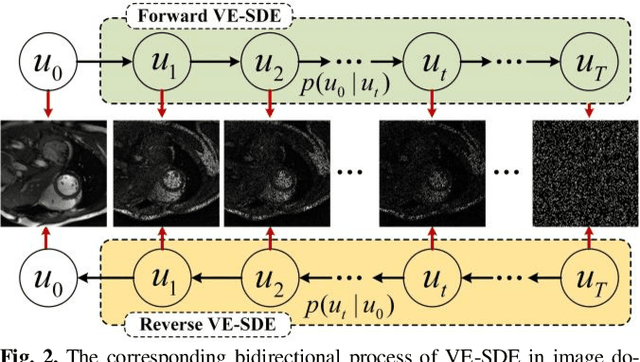
Abstract:Dynamic magnetic resonance image reconstruction from incomplete k-space data has generated great research interest due to its capability to reduce scan time. Never-theless, the reconstruction problem is still challenging due to its ill-posed nature. Recently, diffusion models espe-cially score-based generative models have exhibited great potential in algorithm robustness and usage flexi-bility. Moreover, the unified framework through the variance exploding stochastic differential equation (VE-SDE) is proposed to enable new sampling methods and further extend the capabilities of score-based gener-ative models. Therefore, by taking advantage of the uni-fied framework, we proposed a k-space and image Du-al-Domain collaborative Universal Generative Model (DD-UGM) which combines the score-based prior with low-rank regularization penalty to reconstruct highly under-sampled measurements. More precisely, we extract prior components from both image and k-space domains via a universal generative model and adaptively handle these prior components for faster processing while maintaining good generation quality. Experimental comparisons demonstrated the noise reduction and detail preservation abilities of the proposed method. Much more than that, DD-UGM can reconstruct data of differ-ent frames by only training a single frame image, which reflects the flexibility of the proposed model.
Equilibrated Zeroth-Order Unrolled Deep Networks for Accelerated MRI
Dec 23, 2021
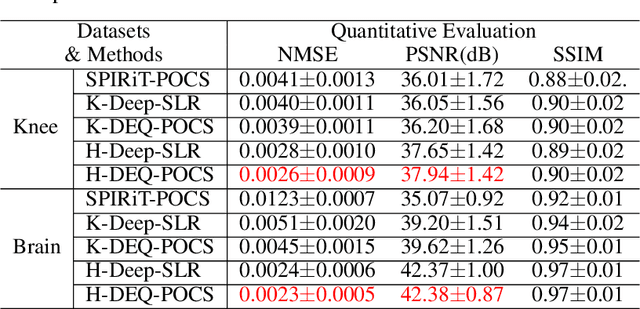

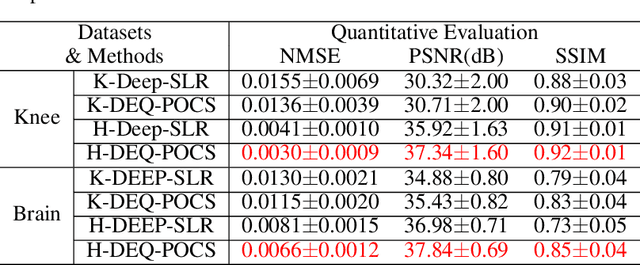
Abstract:Recently, model-driven deep learning unrolls a certain iterative algorithm of a regularization model into a cascade network by replacing the first-order information (i.e., (sub)gradient or proximal operator) of the regularizer with a network module, which appears more explainable and predictable compared to common data-driven networks. Conversely, in theory, there is not necessarily such a functional regularizer whose first-order information matches the replaced network module, which means the network output may not be covered by the original regularization model. Moreover, up to now, there is also no theory to guarantee the global convergence and robustness (regularity) of unrolled networks under realistic assumptions. To bridge this gap, this paper propose to present a safeguarded methodology on network unrolling. Specifically, focusing on accelerated MRI, we unroll a zeroth-order algorithm, of which the network module represents the regularizer itself, so that the network output can be still covered by the regularization model. Furthermore, inspired by the ideal of deep equilibrium models, before backpropagating, we carry out the unrolled iterative network to converge to a fixed point to ensure the convergence. In case the measurement data contains noise, we prove that the proposed network is robust against noisy interference. Finally, numerical experiments show that the proposed network consistently outperforms the state-of-the-art MRI reconstruction methods including traditional regularization methods and other deep learning methods.
SRR-Net: A Super-Resolution-Involved Reconstruction Method for High Resolution MR Imaging
Apr 13, 2021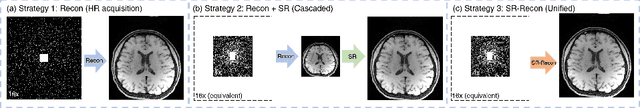
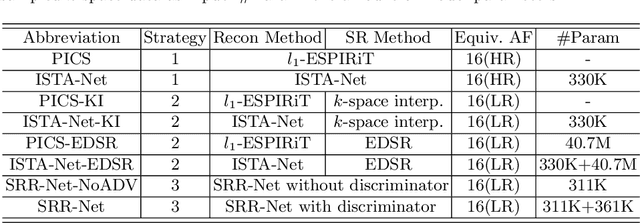
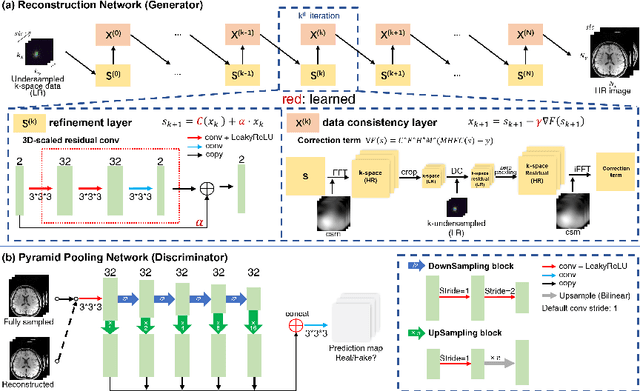
Abstract:Improving the image resolution and acquisition speed of magnetic resonance imaging (MRI) is a challenging problem. There are mainly two strategies dealing with the speed-resolution trade-off: (1) $k$-space undersampling with high-resolution acquisition, and (2) a pipeline of lower resolution image reconstruction and image super-resolution. However, these approaches either have limited performance at certain high acceleration factor or suffer from the error accumulation of two-step structure. In this paper, we combine the idea of MR reconstruction and image super-resolution, and work on recovering HR images from low-resolution under-sampled $k$-space data directly. Particularly, the SR-involved reconstruction can be formulated as a variational problem, and a learnable network unrolled from its solution algorithm is proposed. A discriminator was introduced to enhance the detail refining performance. Experiment results using in-vivo HR multi-coil brain data indicate that the proposed SRR-Net is capable of recovering high-resolution brain images with both good visual quality and perceptual quality.
Deep Manifold Learning for Dynamic MR Imaging
Mar 09, 2021
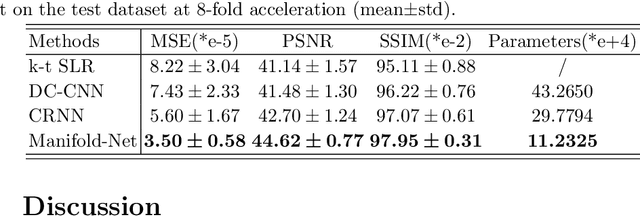
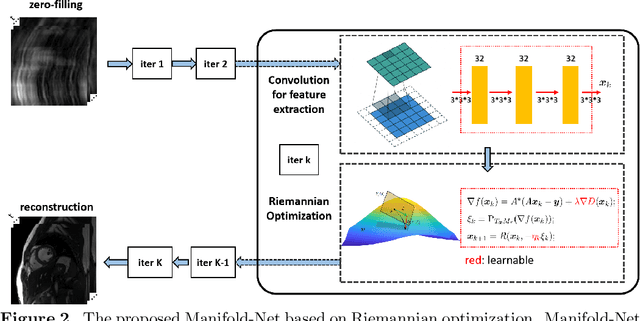

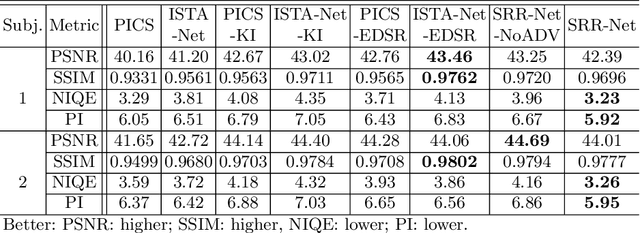
Abstract:Purpose: To develop a deep learning method on a nonlinear manifold to explore the temporal redundancy of dynamic signals to reconstruct cardiac MRI data from highly undersampled measurements. Methods: Cardiac MR image reconstruction is modeled as general compressed sensing (CS) based optimization on a low-rank tensor manifold. The nonlinear manifold is designed to characterize the temporal correlation of dynamic signals. Iterative procedures can be obtained by solving the optimization model on the manifold, including gradient calculation, projection of the gradient to tangent space, and retraction of the tangent space to the manifold. The iterative procedures on the manifold are unrolled to a neural network, dubbed as Manifold-Net. The Manifold-Net is trained using in vivo data with a retrospective electrocardiogram (ECG)-gated segmented bSSFP sequence. Results: Experimental results at high accelerations demonstrate that the proposed method can obtain improved reconstruction compared with a compressed sensing (CS) method k-t SLR and two state-of-the-art deep learning-based methods, DC-CNN and CRNN. Conclusion: This work represents the first study unrolling the optimization on manifolds into neural networks. Specifically, the designed low-rank manifold provides a new technical route for applying low-rank priors in dynamic MR imaging.
Deep Low-rank plus Sparse Network for Dynamic MR Imaging
Oct 26, 2020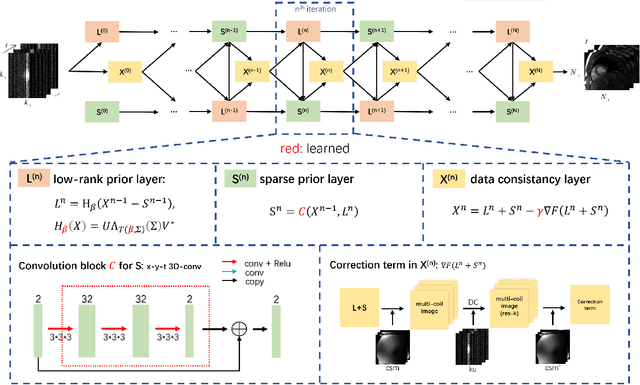
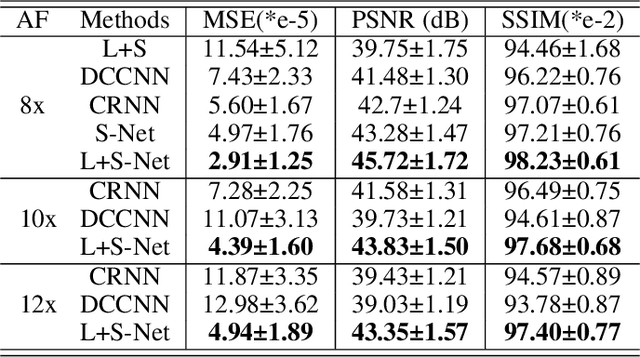
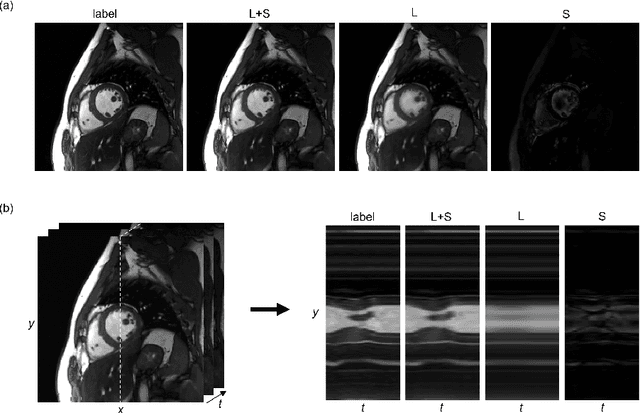
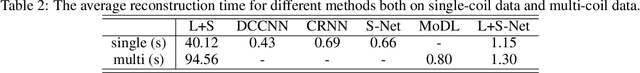
Abstract:In dynamic MR imaging, L+S decomposition, or robust PCA equivalently, has achieved stunning performance. However, the selection of parameters of L+S is empirical, and the acceleration rate is limited, which are the common failings of iterative CS-MRI reconstruction methods. Many deep learning approaches were proposed to address these issues, but few of them used the low-rank prior. In this paper, a model-based low-rank plus sparse network, dubbed as L+S-Net, is proposed for dynamic MR reconstruction. In particular, we use an alternating linearized minimization method to solve the optimization problem with low-rank and sparse regularization. A learned soft singular value thresholding is introduced to make sure the clear separation of L component and S component. Then the iterative steps is unrolled into a network whose regularization parameters are learnable. Experiments on retrospective and prospective cardiac cine dataset show that the proposed model outperforms the state-of-the-art CS and existing deep learning methods.
Deep Low-rank Prior in Dynamic MR Imaging
Jul 06, 2020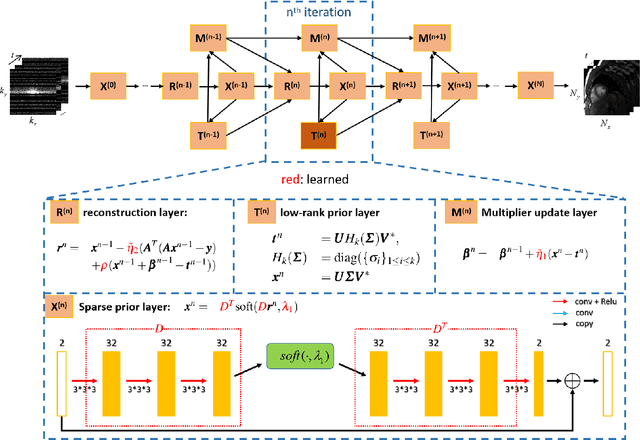
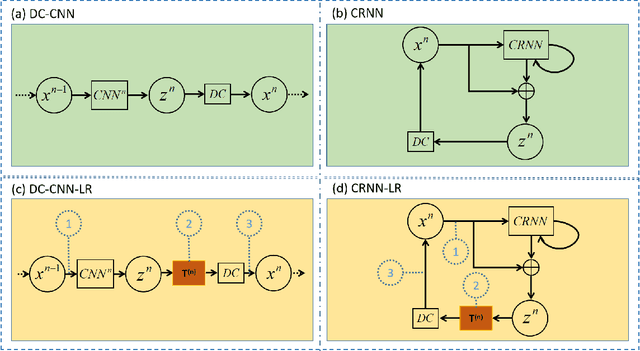
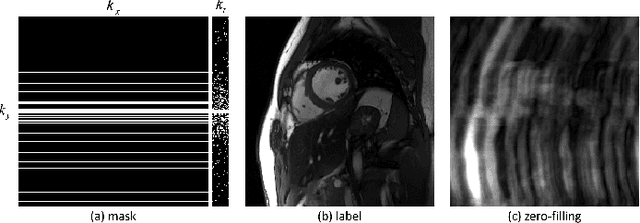
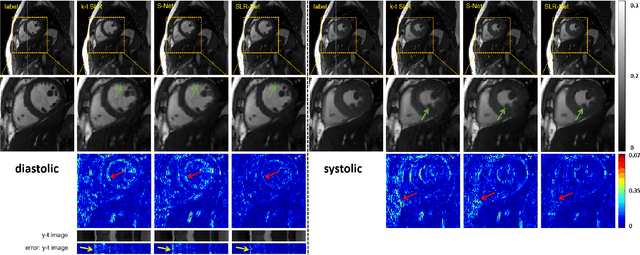
Abstract:The deep learning methods have achieved attractive results in dynamic MR imaging. However, all of these methods only utilize the sparse prior of MR images, while the important low-rank (LR) prior of dynamic MR images is not explored, which limits further improvements of dynamic MR reconstruction. In this paper, a learned singular value thresholding (Learned-SVT) operation is proposed to explore deep low-rank prior in dynamic MR imaging to obtain improved reconstruction results. In particular, we propose two novel and distinct schemes to introduce the learnable low-rank prior into deep network architectures in an unrolling manner and a plug-and-play manner respectively. In the unrolling manner, we propose a model-based unrolling sparse and low-rank network for dynamic MR imaging, dubbed SLR-Net. The SLR-Net is defined over a deep network flow graphs, which is unrolled from the iterative procedures in Iterative Shrinkage-Thresholding Algorithm (ISTA) for optimizing a sparse and low-rank based dynamic MRI model. In the plug-and-play manner, we propose a plug-and-play LR network module that can be easily embedded into any other dynamic MR neural networks without changing the network paradigm. To the best of our knowlegde, this is the first time that a deep low-rank prior has been applied in dynamic MR imaging. Experimental results show that both of the two schemes can further improve the reconstruction results, no matter qualitatively and quantitatively.
An Unsupervised Deep Learning Method for Parallel Cardiac MRI via Time-Interleaved Sampling
Dec 20, 2019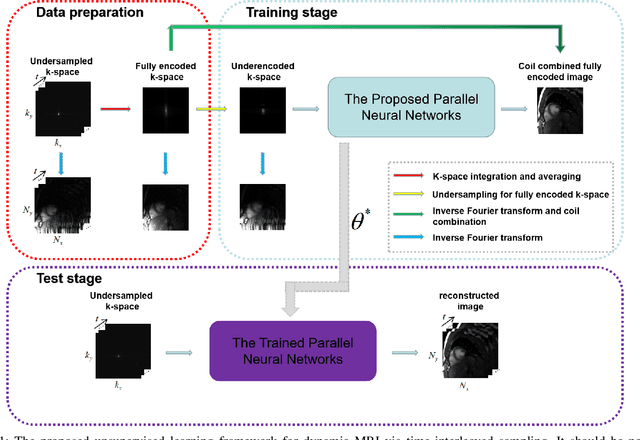
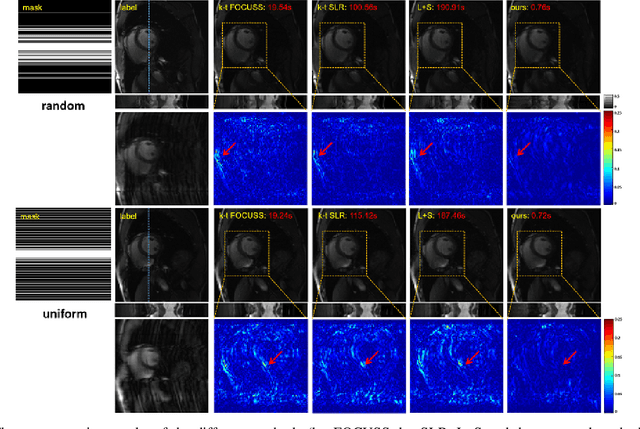
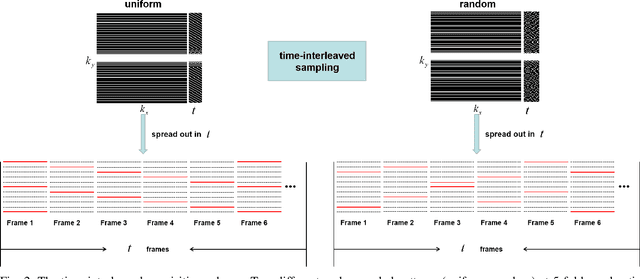
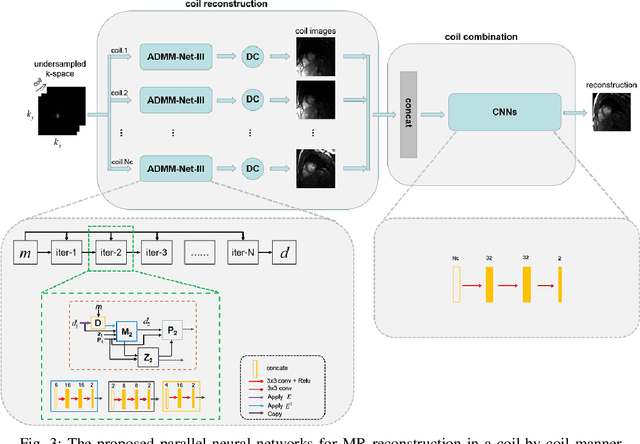
Abstract:Deep learning has achieved good success in cardiac magnetic resonance imaging (MRI) reconstruction, in which convolutional neural networks (CNNs) learn the mapping from undersampled k-space to fully sampled images. Although these deep learning methods can improve reconstruction quality without complex parameter selection or a lengthy reconstruction time compared with iterative methods, the following issues still need to be addressed: 1) all of these methods are based on big data and require a large amount of fully sampled MRI data, which is always difficult for cardiac MRI; 2) All of these methods are only applicable for single-channel images without exploring coil correlation. In this paper, we propose an unsupervised deep learning method for parallel cardiac MRI via a time-interleaved sampling strategy. Specifically, a time-interleaved acquisition scheme is developed to build a set of fully encoded reference data by directly merging the k-space data of adjacent time frames. Then these fully encoded data can be used to train a parallel network for reconstructing images of each coil separately. Finally, the images from each coil are combined together via a CNN to implicitly explore the correlations between coils. The comparisons with classic k-t FOCUSS, k-t SLR and L+S methods on in vivo datasets show that our method can achieve improved reconstruction results in an extremely short amount of time.
LANTERN: learn analysis transform network for dynamic magnetic resonance imaging with small dataset
Aug 24, 2019



Abstract:This paper proposes to learn analysis transform network for dynamic magnetic resonance imaging (LANTERN) with small dataset. Integrating the strength of CS-MRI and deep learning, the proposed framework is highlighted in three components: (i) The spatial and temporal domains are sparsely constrained by using adaptively trained CNN. (ii) We introduce an end-to-end framework to learn the parameters in LANTERN to solve the difficulty of parameter selection in traditional methods. (iii) Compared to existing deep learning reconstruction methods, our reconstruction accuracy is better when the amount of data is limited. Our model is able to fully exploit the redundancy in spatial and temporal of dynamic MR images. We performed quantitative and qualitative analysis of cardiac datasets at different acceleration factors (2x-11x) and different undersampling modes. In comparison with state-of-the-art methods, extensive experiments show that our method achieves consistent better reconstruction performance on the MRI reconstruction in terms of three quantitative metrics (PSNR, SSIM and HFEN) under different undersamling patterns and acceleration factors.
DeepcomplexMRI: Exploiting deep residual network for fast parallel MR imaging with complex convolution
Jul 29, 2019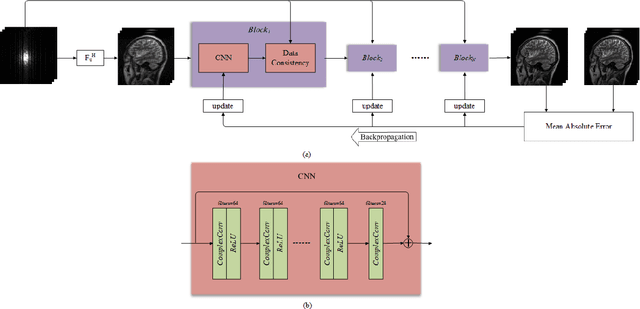
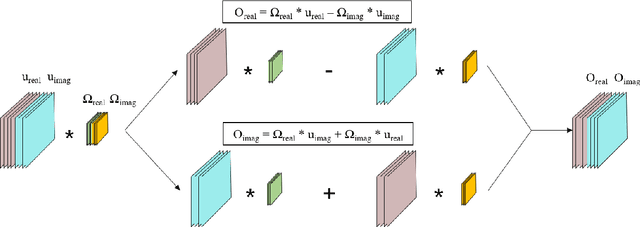

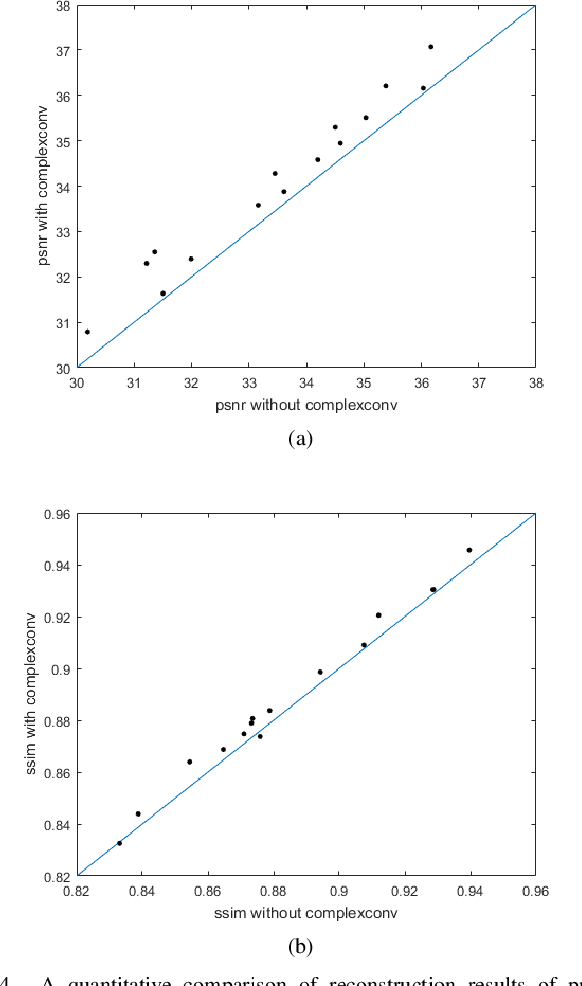
Abstract:This paper proposes a multi-channel image reconstruction method, named DeepcomplexMRI, to accelerate parallel MR imaging with residual complex convolutional neural network. Different from most existing works which rely on the utilization of the coil sensitivities or prior information of predefined transforms, DeepcomplexMRI takes advantage of the availability of a large number of existing multi-channel groudtruth images and uses them as labeled data to train the deep residual convolutional neural network offline. In particular, a complex convolutional network is proposed to take into account the correlation between the real and imaginary parts of MR images. In addition, the k space data consistency is further enforced repeatedly in between layers of the network. The evaluations on in vivo datasets show that the proposed method has the capability to recover the desired multi-channel images. Its comparison with state-of-the-art method also demonstrates that the proposed method can reconstruct the desired MR images more accurately.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge