Yanjie Zhu
Patch-based Reconstruction for Unsupervised Dynamic MRI using Learnable Tensor Function with Implicit Neural Representation
May 28, 2025Abstract:Dynamic MRI plays a vital role in clinical practice by capturing both spatial details and dynamic motion, but its high spatiotemporal resolution is often limited by long scan times. Deep learning (DL)-based methods have shown promising performance in accelerating dynamic MRI. However, most existing algorithms rely on large fully-sampled datasets for training, which are difficult to acquire. Recently, implicit neural representation (INR) has emerged as a powerful scan-specific paradigm for accelerated MRI, which models signals as a continuous function over spatiotemporal coordinates. Although this approach achieves efficient continuous modeling of dynamic images and robust reconstruction, it faces challenges in recovering fine details and increasing computational demands for high dimensional data representation. To enhance both efficiency and reconstruction quality, we propose TenF-INR, a novel patch-based unsupervised framework that employs INR to model bases of tensor decomposition, enabling efficient and accurate modeling of dynamic MR images with learnable tensor functions. By exploiting strong correlations in similar spatial image patches and in the temporal direction, TenF-INR enforces multidimensional low-rankness and implements patch-based reconstruction with the benefits of continuous modeling. We compare TenF-INR with state-of-the-art methods, including supervised DL methods and unsupervised approaches. Experimental results demonstrate that TenF-INR achieves high acceleration factors up to 21, outperforming all comparison methods in image quality, temporal fidelity, and quantitative metrics, even surpassing the supervised methods.
Guided MRI Reconstruction via Schrödinger Bridge
Nov 21, 2024



Abstract:Magnetic Resonance Imaging (MRI) is a multi-contrast imaging technique in which different contrast images share similar structural information. However, conventional diffusion models struggle to effectively leverage this structural similarity. Recently, the Schr\"odinger Bridge (SB), a nonlinear extension of the diffusion model, has been proposed to establish diffusion paths between any distributions, allowing the incorporation of guided priors. This study proposes an SB-based, multi-contrast image-guided reconstruction framework that establishes a diffusion bridge between the guiding and target image distributions. By using the guiding image along with data consistency during sampling, the target image is reconstructed more accurately. To better address structural differences between images, we introduce an inversion strategy from the field of image editing, termed $\mathbf{I}^2$SB-inversion. Experiments on a paried T1 and T2-FLAIR datasets demonstrate that $\mathbf{I}^2$SB-inversion achieve a high acceleration up to 14.4 and outperforms existing methods in terms of both reconstruction accuracy and stability.
RS-MOCO: A deep learning-based topology-preserving image registration method for cardiac T1 mapping
Oct 15, 2024



Abstract:Cardiac T1 mapping can evaluate various clinical symptoms of myocardial tissue. However, there is currently a lack of effective, robust, and efficient methods for motion correction in cardiac T1 mapping. In this paper, we propose a deep learning-based and topology-preserving image registration framework for motion correction in cardiac T1 mapping. Notably, our proposed implicit consistency constraint dubbed BLOC, to some extent preserves the image topology in registration by bidirectional consistency constraint and local anti-folding constraint. To address the contrast variation issue, we introduce a weighted image similarity metric for multimodal registration of cardiac T1-weighted images. Besides, a semi-supervised myocardium segmentation network and a dual-domain attention module are integrated into the framework to further improve the performance of the registration. Numerous comparative experiments, as well as ablation studies, demonstrated the effectiveness and high robustness of our method. The results also indicate that the proposed weighted image similarity metric, specifically crafted for our network, contributes a lot to the enhancement of the motion correction efficacy, while the bidirectional consistency constraint combined with the local anti-folding constraint ensures a more desirable topology-preserving registration mapping.
Quantum Neural Network for Accelerated Magnetic Resonance Imaging
Oct 12, 2024



Abstract:Magnetic resonance image reconstruction starting from undersampled k-space data requires the recovery of many potential nonlinear features, which is very difficult for algorithms to recover these features. In recent years, the development of quantum computing has discovered that quantum convolution can improve network accuracy, possibly due to potential quantum advantages. This article proposes a hybrid neural network containing quantum and classical networks for fast magnetic resonance imaging, and conducts experiments on a quantum computer simulation system. The experimental results indicate that the hybrid network has achieved excellent reconstruction results, and also confirm the feasibility of applying hybrid quantum-classical neural networks into the image reconstruction of rapid magnetic resonance imaging.
Optimized Magnetic Resonance Fingerprinting Using Ziv-Zakai Bound
Oct 10, 2024



Abstract:Magnetic Resonance Fingerprinting (MRF) has emerged as a promising quantitative imaging technique within the field of Magnetic Resonance Imaging (MRI), offers comprehensive insights into tissue properties by simultaneously acquiring multiple tissue parameter maps in a single acquisition. Sequence optimization is crucial for improving the accuracy and efficiency of MRF. In this work, a novel framework for MRF sequence optimization is proposed based on the Ziv-Zakai bound (ZZB). Unlike the Cram\'er-Rao bound (CRB), which aims to enhance the quality of a single fingerprint signal with deterministic parameters, ZZB provides insights into evaluating the minimum mismatch probability for pairs of fingerprint signals within the specified parameter range in MRF. Specifically, the explicit ZZB is derived to establish a lower bound for the discrimination error in the fingerprint signal matching process within MRF. This bound illuminates the intrinsic limitations of MRF sequences, thereby fostering a deeper understanding of existing sequence performance. Subsequently, an optimal experiment design problem based on ZZB was formulated to ascertain the optimal scheme of acquisition parameters, maximizing discrimination power of MRF between different tissue types. Preliminary numerical experiments show that the optimized ZZB scheme outperforms both the conventional and CRB schemes in terms of the reconstruction accuracy of multiple parameter maps.
MR Optimized Reconstruction of Simultaneous Multi-Slice Imaging Using Diffusion Model
Aug 21, 2024Abstract:Diffusion model has been successfully applied to MRI reconstruction, including single and multi-coil acquisition of MRI data. Simultaneous multi-slice imaging (SMS), as a method for accelerating MR acquisition, can significantly reduce scanning time, but further optimization of reconstruction results is still possible. In order to optimize the reconstruction of SMS, we proposed a method to use diffusion model based on slice-GRAPPA and SPIRiT method. approach: Specifically, our method characterizes the prior distribution of SMS data by score matching and characterizes the k-space redundant prior between coils and slices based on self-consistency. With the utilization of diffusion model, we achieved better reconstruction results.The application of diffusion model can further reduce the scanning time of MRI without compromising image quality, making it more advantageous for clinical application
* Accepted as ISMRM 2024 Digital Poster 4024
LINEAR: Learning Implicit Neural Representation With Explicit Physical Priors for Accelerated Quantitative T1rho Mapping
Jul 08, 2024



Abstract:Quantitative T1rho parameter mapping has shown promise in clinical and research studies. However, it suffers from long scan times. Deep learning-based techniques have been successfully applied in accelerated quantitative MR parameter mapping. However, most methods require fully-sampled training dataset, which is impractical in the clinic. In this study, a novel subject-specific unsupervised method based on the implicit neural representation is proposed to reconstruct images from highly undersampled k-space data and estimate parameter maps from reconstructions, which only takes spatiotemporal coordinates as the input. Specifically, the proposed method learned a implicit neural representation of the MR images driven by two explicit priors of images (or k-space data), including the low-rankness of Hankel matrix, and the self-consistency of k-space data. The ablation experiments show that the proposed method can characterize the physical priors of MR images well. Moreover,experimental results of retrospective and prospective data show that the proposed method outperforms the state-of-the-art methods in terms of supressing artifacts and achieving the lowest error.
Diff-DTI: Fast Diffusion Tensor Imaging Using A Feature-Enhanced Joint Diffusion Model
May 24, 2024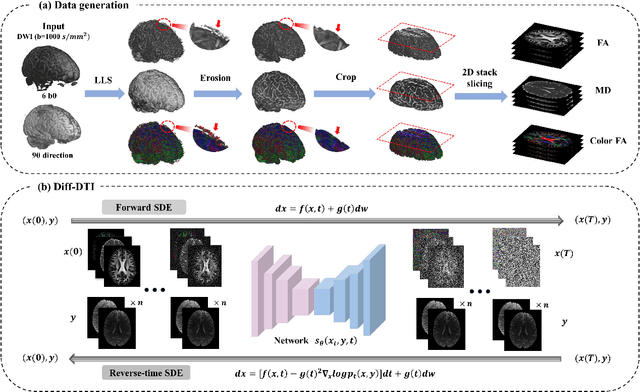
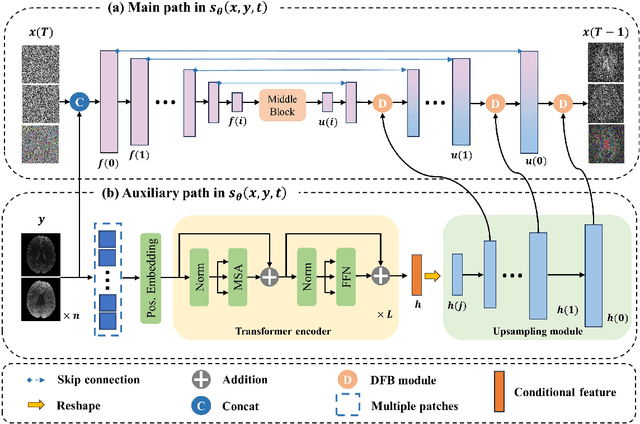
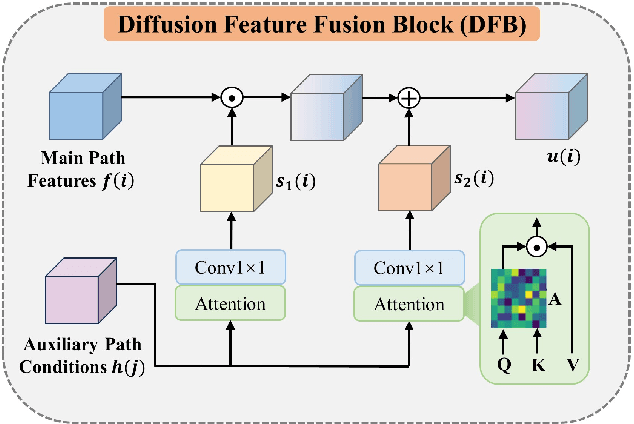
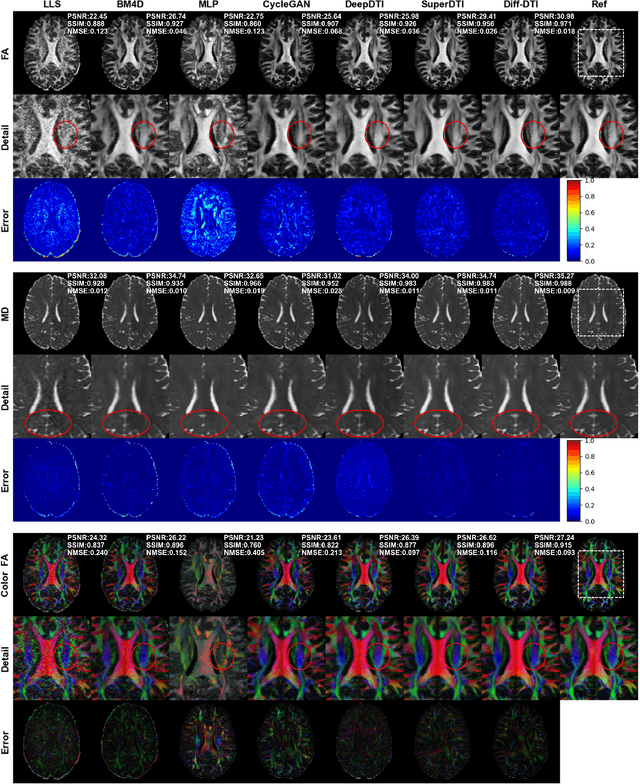
Abstract:Magnetic resonance diffusion tensor imaging (DTI) is a critical tool for neural disease diagnosis. However, long scan time greatly hinders the widespread clinical use of DTI. To accelerate image acquisition, a feature-enhanced joint diffusion model (Diff-DTI) is proposed to obtain accurate DTI parameter maps from a limited number of diffusion-weighted images (DWIs). Diff-DTI introduces a joint diffusion model that directly learns the joint probability distribution of DWIs with DTI parametric maps for conditional generation. Additionally, a feature enhancement fusion mechanism (FEFM) is designed and incorporated into the generative process of Diff-DTI to preserve fine structures in the generated DTI maps. A comprehensive evaluation of the performance of Diff-DTI was conducted on the Human Connectome Project dataset. The results demonstrate that Diff-DTI outperforms existing state-of-the-art fast DTI imaging methods in terms of visual quality and quantitative metrics. Furthermore, Diff-DTI has shown the ability to produce high-fidelity DTI maps with only three DWIs, thus overcoming the requirement of a minimum of six DWIs for DTI.
Joint Diffusion: Mutual Consistency-Driven Diffusion Model for PET-MRI Co-Reconstruction
Nov 24, 2023Abstract:Positron Emission Tomography and Magnetic Resonance Imaging (PET-MRI) systems can obtain functional and anatomical scans. PET suffers from a low signal-to-noise ratio. Meanwhile, the k-space data acquisition process in MRI is time-consuming. The study aims to accelerate MRI and enhance PET image quality. Conventional approaches involve the separate reconstruction of each modality within PET-MRI systems. However, there exists complementary information among multi-modal images. The complementary information can contribute to image reconstruction. In this study, we propose a novel PET-MRI joint reconstruction model employing a mutual consistency-driven diffusion mode, namely MC-Diffusion. MC-Diffusion learns the joint probability distribution of PET and MRI for utilizing complementary information. We conducted a series of contrast experiments about LPLS, Joint ISAT-net and MC-Diffusion by the ADNI dataset. The results underscore the qualitative and quantitative improvements achieved by MC-Diffusion, surpassing the state-of-the-art method.
A Two-Stage Generative Model with CycleGAN and Joint Diffusion for MRI-based Brain Tumor Detection
Nov 06, 2023Abstract:Accurate detection and segmentation of brain tumors is critical for medical diagnosis. However, current supervised learning methods require extensively annotated images and the state-of-the-art generative models used in unsupervised methods often have limitations in covering the whole data distribution. In this paper, we propose a novel framework Two-Stage Generative Model (TSGM) that combines Cycle Generative Adversarial Network (CycleGAN) and Variance Exploding stochastic differential equation using joint probability (VE-JP) to improve brain tumor detection and segmentation. The CycleGAN is trained on unpaired data to generate abnormal images from healthy images as data prior. Then VE-JP is implemented to reconstruct healthy images using synthetic paired abnormal images as a guide, which alters only pathological regions but not regions of healthy. Notably, our method directly learned the joint probability distribution for conditional generation. The residual between input and reconstructed images suggests the abnormalities and a thresholding method is subsequently applied to obtain segmentation results. Furthermore, the multimodal results are weighted with different weights to improve the segmentation accuracy further. We validated our method on three datasets, and compared with other unsupervised methods for anomaly detection and segmentation. The DSC score of 0.8590 in BraTs2020 dataset, 0.6226 in ITCS dataset and 0.7403 in In-house dataset show that our method achieves better segmentation performance and has better generalization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge