Chentao Cao
From Debate to Equilibrium: Belief-Driven Multi-Agent LLM Reasoning via Bayesian Nash Equilibrium
Jun 09, 2025Abstract:Multi-agent frameworks can substantially boost the reasoning power of large language models (LLMs), but they typically incur heavy computational costs and lack convergence guarantees. To overcome these challenges, we recast multi-LLM coordination as an incomplete-information game and seek a Bayesian Nash equilibrium (BNE), in which each agent optimally responds to its probabilistic beliefs about the strategies of others. We introduce Efficient Coordination via Nash Equilibrium (ECON), a hierarchical reinforcement-learning paradigm that marries distributed reasoning with centralized final output. Under ECON, each LLM independently selects responses that maximize its expected reward, conditioned on its beliefs about co-agents, without requiring costly inter-agent exchanges. We mathematically prove that ECON attains a markedly tighter regret bound than non-equilibrium multi-agent schemes. Empirically, ECON outperforms existing multi-LLM approaches by 11.2% on average across six benchmarks spanning complex reasoning and planning tasks. Further experiments demonstrate ECON's ability to flexibly incorporate additional models, confirming its scalability and paving the way toward larger, more powerful multi-LLM ensembles. The code is publicly available at: https://github.com/tmlr-group/ECON.
Noisy Test-Time Adaptation in Vision-Language Models
Feb 20, 2025
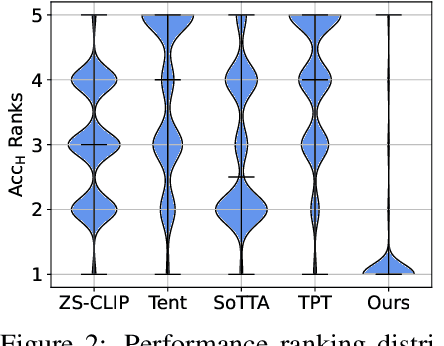
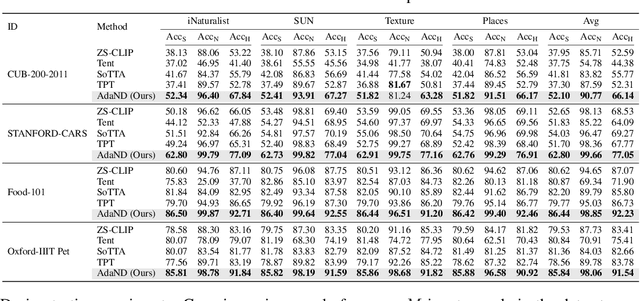
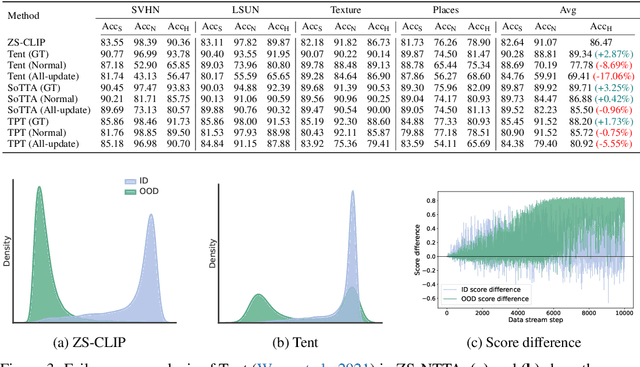
Abstract:Test-time adaptation (TTA) aims to address distribution shifts between source and target data by relying solely on target data during testing. In open-world scenarios, models often encounter noisy samples, i.e., samples outside the in-distribution (ID) label space. Leveraging the zero-shot capability of pre-trained vision-language models (VLMs), this paper introduces Zero-Shot Noisy TTA (ZS-NTTA), focusing on adapting the model to target data with noisy samples during test-time in a zero-shot manner. We find existing TTA methods underperform under ZS-NTTA, often lagging behind even the frozen model. We conduct comprehensive experiments to analyze this phenomenon, revealing that the negative impact of unfiltered noisy data outweighs the benefits of clean data during model updating. Also, adapting a classifier for ID classification and noise detection hampers both sub-tasks. Built on this, we propose a framework that decouples the classifier and detector, focusing on developing an individual detector while keeping the classifier frozen. Technically, we introduce the Adaptive Noise Detector (AdaND), which utilizes the frozen model's outputs as pseudo-labels to train a noise detector. To handle clean data streams, we further inject Gaussian noise during adaptation, preventing the detector from misclassifying clean samples as noisy. Beyond the ZS-NTTA, AdaND can also improve the zero-shot out-of-distribution (ZS-OOD) detection ability of VLMs. Experiments show that AdaND outperforms in both ZS-NTTA and ZS-OOD detection. On ImageNet, AdaND achieves a notable improvement of $8.32\%$ in harmonic mean accuracy ($\text{Acc}_\text{H}$) for ZS-NTTA and $9.40\%$ in FPR95 for ZS-OOD detection, compared to SOTA methods. Importantly, AdaND is computationally efficient and comparable to the model-frozen method. The code is publicly available at: https://github.com/tmlr-group/ZS-NTTA.
Envisioning Outlier Exposure by Large Language Models for Out-of-Distribution Detection
Jun 02, 2024Abstract:Detecting out-of-distribution (OOD) samples is essential when deploying machine learning models in open-world scenarios. Zero-shot OOD detection, requiring no training on in-distribution (ID) data, has been possible with the advent of vision-language models like CLIP. Existing methods build a text-based classifier with only closed-set labels. However, this largely restricts the inherent capability of CLIP to recognize samples from large and open label space. In this paper, we propose to tackle this constraint by leveraging the expert knowledge and reasoning capability of large language models (LLM) to Envision potential Outlier Exposure, termed EOE, without access to any actual OOD data. Owing to better adaptation to open-world scenarios, EOE can be generalized to different tasks, including far, near, and fine-grained OOD detection. Technically, we design (1) LLM prompts based on visual similarity to generate potential outlier class labels specialized for OOD detection, as well as (2) a new score function based on potential outlier penalty to distinguish hard OOD samples effectively. Empirically, EOE achieves state-of-the-art performance across different OOD tasks and can be effectively scaled to the ImageNet-1K dataset. The code is publicly available at: https://github.com/tmlr-group/EOE.
A Two-Stage Generative Model with CycleGAN and Joint Diffusion for MRI-based Brain Tumor Detection
Nov 06, 2023Abstract:Accurate detection and segmentation of brain tumors is critical for medical diagnosis. However, current supervised learning methods require extensively annotated images and the state-of-the-art generative models used in unsupervised methods often have limitations in covering the whole data distribution. In this paper, we propose a novel framework Two-Stage Generative Model (TSGM) that combines Cycle Generative Adversarial Network (CycleGAN) and Variance Exploding stochastic differential equation using joint probability (VE-JP) to improve brain tumor detection and segmentation. The CycleGAN is trained on unpaired data to generate abnormal images from healthy images as data prior. Then VE-JP is implemented to reconstruct healthy images using synthetic paired abnormal images as a guide, which alters only pathological regions but not regions of healthy. Notably, our method directly learned the joint probability distribution for conditional generation. The residual between input and reconstructed images suggests the abnormalities and a thresholding method is subsequently applied to obtain segmentation results. Furthermore, the multimodal results are weighted with different weights to improve the segmentation accuracy further. We validated our method on three datasets, and compared with other unsupervised methods for anomaly detection and segmentation. The DSC score of 0.8590 in BraTs2020 dataset, 0.6226 in ITCS dataset and 0.7403 in In-house dataset show that our method achieves better segmentation performance and has better generalization.
Physics-Informed DeepMRI: Bridging the Gap from Heat Diffusion to k-Space Interpolation
Aug 30, 2023



Abstract:In the field of parallel imaging (PI), alongside image-domain regularization methods, substantial research has been dedicated to exploring $k$-space interpolation. However, the interpretability of these methods remains an unresolved issue. Furthermore, these approaches currently face acceleration limitations that are comparable to those experienced by image-domain methods. In order to enhance interpretability and overcome the acceleration limitations, this paper introduces an interpretable framework that unifies both $k$-space interpolation techniques and image-domain methods, grounded in the physical principles of heat diffusion equations. Building upon this foundational framework, a novel $k$-space interpolation method is proposed. Specifically, we model the process of high-frequency information attenuation in $k$-space as a heat diffusion equation, while the effort to reconstruct high-frequency information from low-frequency regions can be conceptualized as a reverse heat equation. However, solving the reverse heat equation poses a challenging inverse problem. To tackle this challenge, we modify the heat equation to align with the principles of magnetic resonance PI physics and employ the score-based generative method to precisely execute the modified reverse heat diffusion. Finally, experimental validation conducted on publicly available datasets demonstrates the superiority of the proposed approach over traditional $k$-space interpolation methods, deep learning-based $k$-space interpolation methods, and conventional diffusion models in terms of reconstruction accuracy, particularly in high-frequency regions.
Synthesizing PET images from High-field and Ultra-high-field MR images Using Joint Diffusion Attention Model
May 06, 2023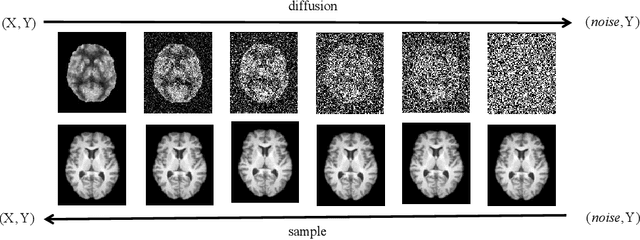
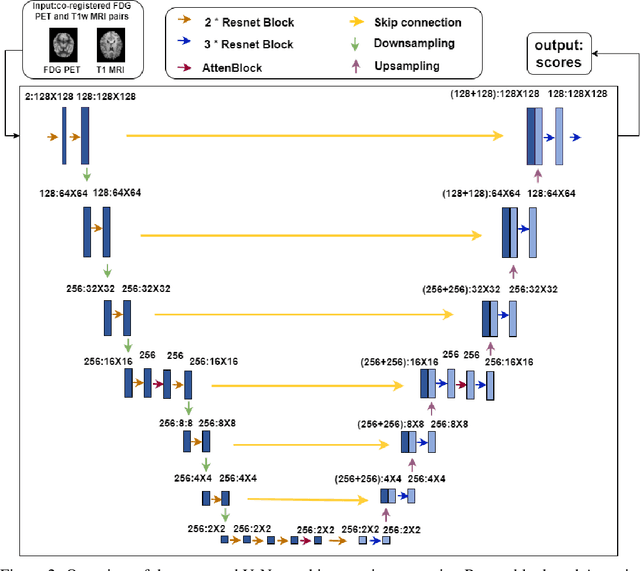
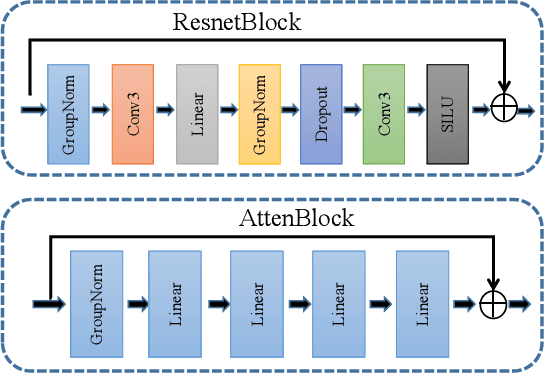
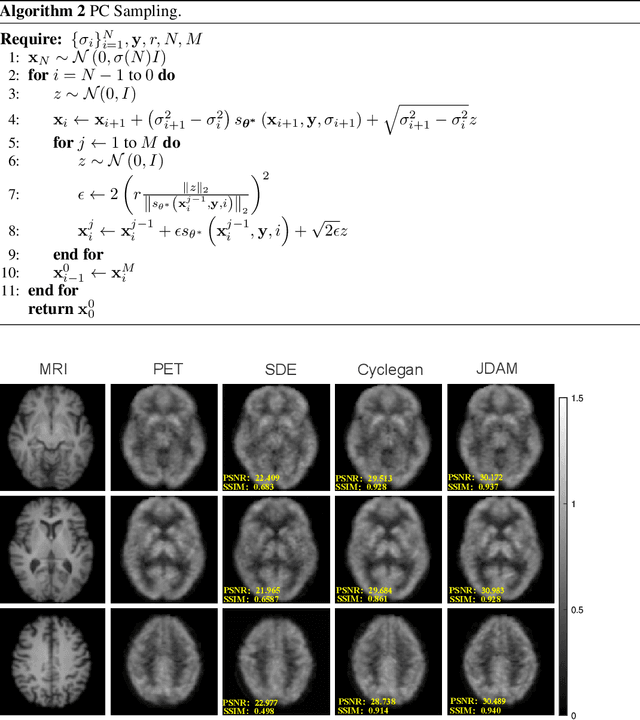
Abstract:MRI and PET are crucial diagnostic tools for brain diseases, as they provide complementary information on brain structure and function. However, PET scanning is costly and involves radioactive exposure, resulting in a lack of PET. Moreover, simultaneous PET and MRI at ultra-high-field are currently hardly infeasible. Ultra-high-field imaging has unquestionably proven valuable in both clinical and academic settings, especially in the field of cognitive neuroimaging. These motivate us to propose a method for synthetic PET from high-filed MRI and ultra-high-field MRI. From a statistical perspective, the joint probability distribution (JPD) is the most direct and fundamental means of portraying the correlation between PET and MRI. This paper proposes a novel joint diffusion attention model which has the joint probability distribution and attention strategy, named JDAM. JDAM has a diffusion process and a sampling process. The diffusion process involves the gradual diffusion of PET to Gaussian noise by adding Gaussian noise, while MRI remains fixed. JPD of MRI and noise-added PET was learned in the diffusion process. The sampling process is a predictor-corrector. PET images were generated from MRI by JPD of MRI and noise-added PET. The predictor is a reverse diffusion process and the corrector is Langevin dynamics. Experimental results on the public Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset demonstrate that the proposed method outperforms state-of-the-art CycleGAN for high-field MRI (3T MRI). Finally, synthetic PET images from the ultra-high-field (5T MRI and 7T MRI) be attempted, providing a possibility for ultra-high-field PET-MRI imaging.
Meta-Learning Enabled Score-Based Generative Model for 1.5T-Like Image Reconstruction from 0.5T MRI
May 04, 2023



Abstract:Magnetic resonance imaging (MRI) is known to have reduced signal-to-noise ratios (SNR) at lower field strengths, leading to signal degradation when producing a low-field MRI image from a high-field one. Therefore, reconstructing a high-field-like image from a low-field MRI is a complex problem due to the ill-posed nature of the task. Additionally, obtaining paired low-field and high-field MR images is often not practical. We theoretically uncovered that the combination of these challenges renders conventional deep learning methods that directly learn the mapping from a low-field MR image to a high-field MR image unsuitable. To overcome these challenges, we introduce a novel meta-learning approach that employs a teacher-student mechanism. Firstly, an optimal-transport-driven teacher learns the degradation process from high-field to low-field MR images and generates pseudo-paired high-field and low-field MRI images. Then, a score-based student solves the inverse problem of reconstructing a high-field-like MR image from a low-field MRI within the framework of iterative regularization, by learning the joint distribution of pseudo-paired images to act as a regularizer. Experimental results on real low-field MRI data demonstrate that our proposed method outperforms state-of-the-art unpaired learning methods.
SPIRiT-Diffusion: Self-Consistency Driven Diffusion Model for Accelerated MRI
Apr 11, 2023



Abstract:Diffusion models are a leading method for image generation and have been successfully applied in magnetic resonance imaging (MRI) reconstruction. Current diffusion-based reconstruction methods rely on coil sensitivity maps (CSM) to reconstruct multi-coil data. However, it is difficult to accurately estimate CSMs in practice use, resulting in degradation of the reconstruction quality. To address this issue, we propose a self-consistency-driven diffusion model inspired by the iterative self-consistent parallel imaging (SPIRiT), namely SPIRiT-Diffusion. Specifically, the iterative solver of the self-consistent term in SPIRiT is utilized to design a novel stochastic differential equation (SDE) for diffusion process. Then $\textit{k}$-space data can be interpolated directly during the reverse diffusion process, instead of using CSM to separate and combine individual coil images. This method indicates that the optimization model can be used to design SDE in diffusion models, driving the diffusion process strongly conforming with the physics involved in the optimization model, dubbed model-driven diffusion. The proposed SPIRiT-Diffusion method was evaluated on a 3D joint Intracranial and Carotid Vessel Wall imaging dataset. The results demonstrate that it outperforms the CSM-based reconstruction methods, and achieves high reconstruction quality at a high acceleration rate of 10.
SPIRiT-Diffusion: SPIRiT-driven Score-Based Generative Modeling for Vessel Wall imaging
Dec 14, 2022

Abstract:Diffusion model is the most advanced method in image generation and has been successfully applied to MRI reconstruction. However, the existing methods do not consider the characteristics of multi-coil acquisition of MRI data. Therefore, we give a new diffusion model, called SPIRiT-Diffusion, based on the SPIRiT iterative reconstruction algorithm. Specifically, SPIRiT-Diffusion characterizes the prior distribution of coil-by-coil images by score matching and characterizes the k-space redundant prior between coils based on self-consistency. With sufficient prior constraint utilized, we achieve superior reconstruction results on the joint Intracranial and Carotid Vessel Wall imaging dataset.
Self-Score: Self-Supervised Learning on Score-Based Models for MRI Reconstruction
Sep 02, 2022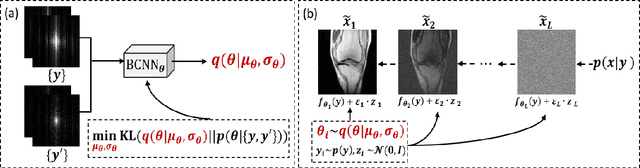
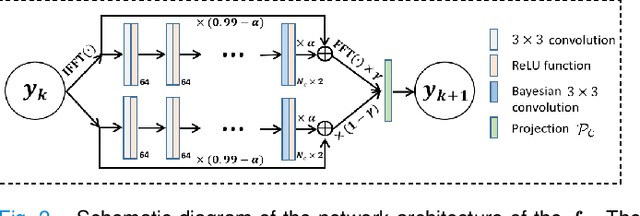

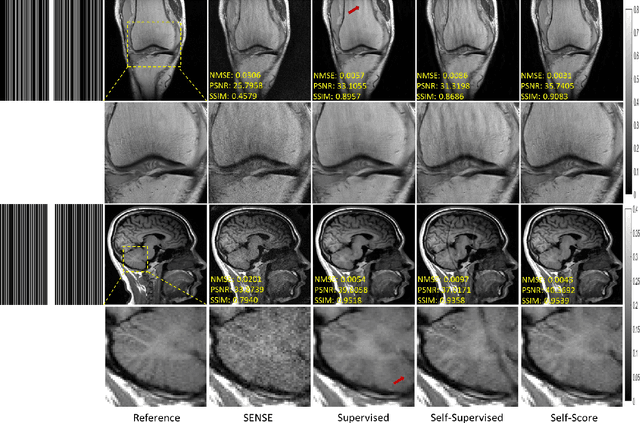
Abstract:Recently, score-based diffusion models have shown satisfactory performance in MRI reconstruction. Most of these methods require a large amount of fully sampled MRI data as a training set, which, sometimes, is difficult to acquire in practice. This paper proposes a fully-sampled-data-free score-based diffusion model for MRI reconstruction, which learns the fully sampled MR image prior in a self-supervised manner on undersampled data. Specifically, we first infer the fully sampled MR image distribution from the undersampled data by Bayesian deep learning, then perturb the data distribution and approximate their probability density gradient by training a score function. Leveraging the learned score function as a prior, we can reconstruct the MR image by performing conditioned Langevin Markov chain Monte Carlo (MCMC) sampling. Experiments on the public dataset show that the proposed method outperforms existing self-supervised MRI reconstruction methods and achieves comparable performances with the conventional (fully sampled data trained) score-based diffusion methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge