Shaonan Liu
Benchmarking Egocentric Clinical Intent Understanding Capability for Medical Multimodal Large Language Models
Jan 11, 2026Abstract:Medical Multimodal Large Language Models (Med-MLLMs) require egocentric clinical intent understanding for real-world deployment, yet existing benchmarks fail to evaluate this critical capability. To address these challenges, we introduce MedGaze-Bench, the first benchmark leveraging clinician gaze as a Cognitive Cursor to assess intent understanding across surgery, emergency simulation, and diagnostic interpretation. Our benchmark addresses three fundamental challenges: visual homogeneity of anatomical structures, strict temporal-causal dependencies in clinical workflows, and implicit adherence to safety protocols. We propose a Three-Dimensional Clinical Intent Framework evaluating: (1) Spatial Intent: discriminating precise targets amid visual noise, (2) Temporal Intent: inferring causal rationale through retrospective and prospective reasoning, and (3) Standard Intent: verifying protocol compliance through safety checks. Beyond accuracy metrics, we introduce Trap QA mechanisms to stress-test clinical reliability by penalizing hallucinations and cognitive sycophancy. Experiments reveal current MLLMs struggle with egocentric intent due to over-reliance on global features, leading to fabricated observations and uncritical acceptance of invalid instructions.
GEM: Context-Aware Gaze EstiMation with Visual Search Behavior Matching for Chest Radiograph
Aug 10, 2024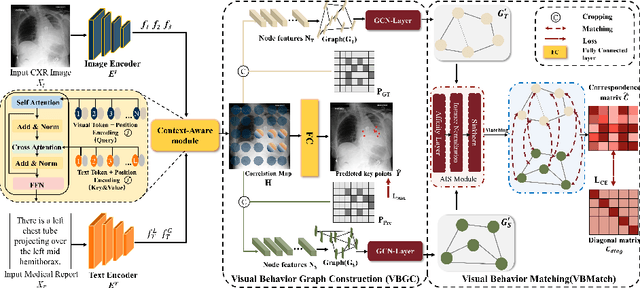
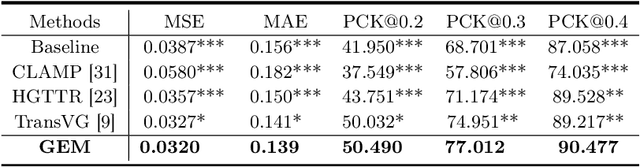
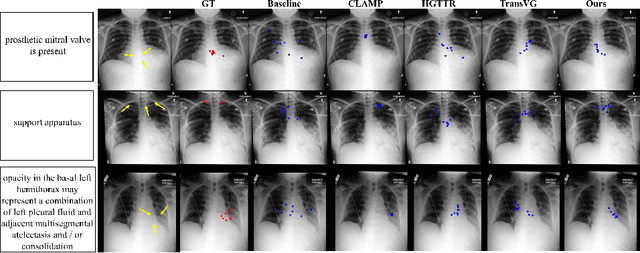
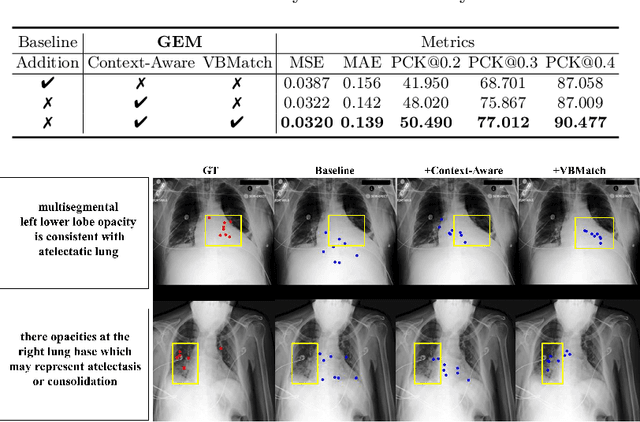
Abstract:Gaze estimation is pivotal in human scene comprehension tasks, particularly in medical diagnostic analysis. Eye-tracking technology facilitates the recording of physicians' ocular movements during image interpretation, thereby elucidating their visual attention patterns and information-processing strategies. In this paper, we initially define the context-aware gaze estimation problem in medical radiology report settings. To understand the attention allocation and cognitive behavior of radiologists during the medical image interpretation process, we propose a context-aware Gaze EstiMation (GEM) network that utilizes eye gaze data collected from radiologists to simulate their visual search behavior patterns throughout the image interpretation process. It consists of a context-awareness module, visual behavior graph construction, and visual behavior matching. Within the context-awareness module, we achieve intricate multimodal registration by establishing connections between medical reports and images. Subsequently, for a more accurate simulation of genuine visual search behavior patterns, we introduce a visual behavior graph structure, capturing such behavior through high-order relationships (edges) between gaze points (nodes). To maintain the authenticity of visual behavior, we devise a visual behavior-matching approach, adjusting the high-order relationships between them by matching the graph constructed from real and estimated gaze points. Extensive experiments on four publicly available datasets demonstrate the superiority of GEM over existing methods and its strong generalizability, which also provides a new direction for the effective utilization of diverse modalities in medical image interpretation and enhances the interpretability of models in the field of medical imaging. https://github.com/Tiger-SN/GEM
Self-Score: Self-Supervised Learning on Score-Based Models for MRI Reconstruction
Sep 02, 2022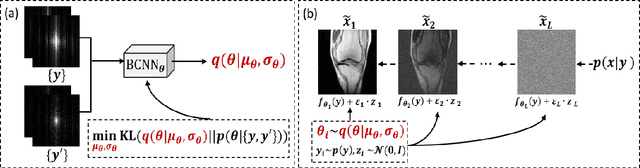
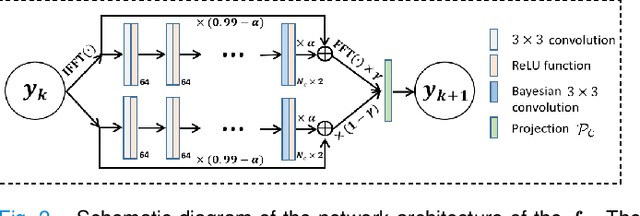

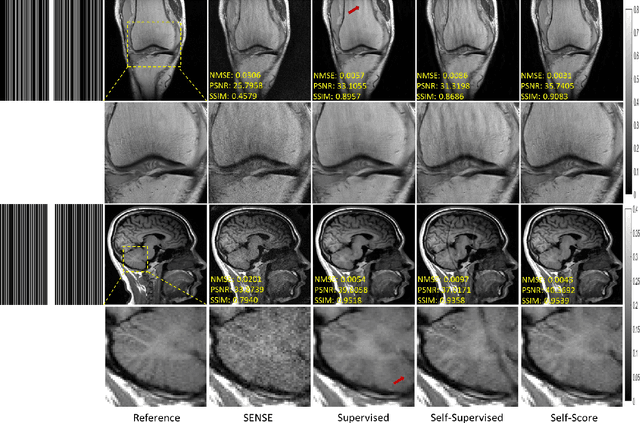
Abstract:Recently, score-based diffusion models have shown satisfactory performance in MRI reconstruction. Most of these methods require a large amount of fully sampled MRI data as a training set, which, sometimes, is difficult to acquire in practice. This paper proposes a fully-sampled-data-free score-based diffusion model for MRI reconstruction, which learns the fully sampled MR image prior in a self-supervised manner on undersampled data. Specifically, we first infer the fully sampled MR image distribution from the undersampled data by Bayesian deep learning, then perturb the data distribution and approximate their probability density gradient by training a score function. Leveraging the learned score function as a prior, we can reconstruct the MR image by performing conditioned Langevin Markov chain Monte Carlo (MCMC) sampling. Experiments on the public dataset show that the proposed method outperforms existing self-supervised MRI reconstruction methods and achieves comparable performances with the conventional (fully sampled data trained) score-based diffusion methods.
High-Frequency Space Diffusion Models for Accelerated MRI
Aug 15, 2022



Abstract:Denoising diffusion probabilistic models (DDPMs) have been shown to have superior performances in MRI reconstruction. From the perspective of continuous stochastic differential equations (SDEs), the reverse process of DDPM can be seen as maximizing the energy of the reconstructed MR image, leading to SDE sequence divergence. For this reason, a modified high-frequency DDPM model is proposed for MRI reconstruction. From its continuous SDE viewpoint, termed high-frequency space SDE (HFS-SDE), the energy concentrated low-frequency part of the MR image is no longer amplified, and the diffusion process focuses more on acquiring high-frequency prior information. It not only improves the stability of the diffusion model but also provides the possibility of better recovery of high-frequency details. Experiments on the publicly fastMRI dataset show that our proposed HFS-SDE outperforms the DDPM-driven VP-SDE, supervised deep learning methods and traditional parallel imaging methods in terms of stability and reconstruction accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge