Xiaoling Luo
Vision KAN: Towards an Attention-Free Backbone for Vision with Kolmogorov-Arnold Networks
Jan 29, 2026Abstract:Attention mechanisms have become a key module in modern vision backbones due to their ability to model long-range dependencies. However, their quadratic complexity in sequence length and the difficulty of interpreting attention weights limit both scalability and clarity. Recent attention-free architectures demonstrate that strong performance can be achieved without pairwise attention, motivating the search for alternatives. In this work, we introduce Vision KAN (ViK), an attention-free backbone inspired by the Kolmogorov-Arnold Networks. At its core lies MultiPatch-RBFKAN, a unified token mixer that combines (a) patch-wise nonlinear transform with Radial Basis Function-based KANs, (b) axis-wise separable mixing for efficient local propagation, and (c) low-rank global mapping for long-range interaction. Employing as a drop-in replacement for attention modules, this formulation tackles the prohibitive cost of full KANs on high-resolution features by adopting a patch-wise grouping strategy with lightweight operators to restore cross-patch dependencies. Experiments on ImageNet-1K show that ViK achieves competitive accuracy with linear complexity, demonstrating the potential of KAN-based token mixing as an efficient and theoretically grounded alternative to attention.
MMedExpert-R1: Strengthening Multimodal Medical Reasoning via Domain-Specific Adaptation and Clinical Guideline Reinforcement
Jan 16, 2026Abstract:Medical Vision-Language Models (MedVLMs) excel at perception tasks but struggle with complex clinical reasoning required in real-world scenarios. While reinforcement learning (RL) has been explored to enhance reasoning capabilities, existing approaches face critical mismatches: the scarcity of deep reasoning data, cold-start limits multi-specialty alignment, and standard RL algorithms fail to model clinical reasoning diversity. We propose MMedExpert-R1, a novel reasoning MedVLM that addresses these challenges through domain-specific adaptation and clinical guideline reinforcement. We construct MMedExpert, a high-quality dataset of 10K samples across four specialties with step-by-step reasoning traces. Our Domain-Specific Adaptation (DSA) creates specialty-specific LoRA modules to provide diverse initialization, while Guideline-Based Advantages (GBA) explicitly models different clinical reasoning perspectives to align with real-world diagnostic strategies. Conflict-Aware Capability Integration then merges these specialized experts into a unified agent, ensuring robust multi-specialty alignment. Comprehensive experiments demonstrate state-of-the-art performance, with our 7B model achieving 27.50 on MedXpert-MM and 83.03 on OmniMedVQA, establishing a robust foundation for reliable multimodal medical reasoning systems.
Benchmarking Egocentric Clinical Intent Understanding Capability for Medical Multimodal Large Language Models
Jan 11, 2026Abstract:Medical Multimodal Large Language Models (Med-MLLMs) require egocentric clinical intent understanding for real-world deployment, yet existing benchmarks fail to evaluate this critical capability. To address these challenges, we introduce MedGaze-Bench, the first benchmark leveraging clinician gaze as a Cognitive Cursor to assess intent understanding across surgery, emergency simulation, and diagnostic interpretation. Our benchmark addresses three fundamental challenges: visual homogeneity of anatomical structures, strict temporal-causal dependencies in clinical workflows, and implicit adherence to safety protocols. We propose a Three-Dimensional Clinical Intent Framework evaluating: (1) Spatial Intent: discriminating precise targets amid visual noise, (2) Temporal Intent: inferring causal rationale through retrospective and prospective reasoning, and (3) Standard Intent: verifying protocol compliance through safety checks. Beyond accuracy metrics, we introduce Trap QA mechanisms to stress-test clinical reliability by penalizing hallucinations and cognitive sycophancy. Experiments reveal current MLLMs struggle with egocentric intent due to over-reliance on global features, leading to fabricated observations and uncritical acceptance of invalid instructions.
Enhancing Multimodal Protein Function Prediction Through Dual-Branch Dynamic Selection with Reconstructive Pre-Training
Nov 06, 2025Abstract:Multimodal protein features play a crucial role in protein function prediction. However, these features encompass a wide range of information, ranging from structural data and sequence features to protein attributes and interaction networks, making it challenging to decipher their complex interconnections. In this work, we propose a multimodal protein function prediction method (DSRPGO) by utilizing dynamic selection and reconstructive pre-training mechanisms. To acquire complex protein information, we introduce reconstructive pre-training to mine more fine-grained information with low semantic levels. Moreover, we put forward the Bidirectional Interaction Module (BInM) to facilitate interactive learning among multimodal features. Additionally, to address the difficulty of hierarchical multi-label classification in this task, a Dynamic Selection Module (DSM) is designed to select the feature representation that is most conducive to current protein function prediction. Our proposed DSRPGO model improves significantly in BPO, MFO, and CCO on human datasets, thereby outperforming other benchmark models.
Wavelet-based Global-Local Interaction Network with Cross-Attention for Multi-View Diabetic Retinopathy Detection
Mar 25, 2025Abstract:Multi-view diabetic retinopathy (DR) detection has recently emerged as a promising method to address the issue of incomplete lesions faced by single-view DR. However, it is still challenging due to the variable sizes and scattered locations of lesions. Furthermore, existing multi-view DR methods typically merge multiple views without considering the correlations and redundancies of lesion information across them. Therefore, we propose a novel method to overcome the challenges of difficult lesion information learning and inadequate multi-view fusion. Specifically, we introduce a two-branch network to obtain both local lesion features and their global dependencies. The high-frequency component of the wavelet transform is used to exploit lesion edge information, which is then enhanced by global semantic to facilitate difficult lesion learning. Additionally, we present a cross-view fusion module to improve multi-view fusion and reduce redundancy. Experimental results on large public datasets demonstrate the effectiveness of our method. The code is open sourced on https://github.com/HuYongting/WGLIN.
HySurvPred: Multimodal Hyperbolic Embedding with Angle-Aware Hierarchical Contrastive Learning and Uncertainty Constraints for Survival Prediction
Mar 18, 2025Abstract:Multimodal learning that integrates histopathology images and genomic data holds great promise for cancer survival prediction. However, existing methods face key limitations: 1) They rely on multimodal mapping and metrics in Euclidean space, which cannot fully capture the hierarchical structures in histopathology (among patches from different resolutions) and genomics data (from genes to pathways). 2) They discretize survival time into independent risk intervals, which ignores its continuous and ordinal nature and fails to achieve effective optimization. 3) They treat censorship as a binary indicator, excluding censored samples from model optimization and not making full use of them. To address these challenges, we propose HySurvPred, a novel framework for survival prediction that integrates three key modules: Multimodal Hyperbolic Mapping (MHM), Angle-aware Ranking-based Contrastive Loss (ARCL) and Censor-Conditioned Uncertainty Constraint (CUC). Instead of relying on Euclidean space, we design the MHM module to explore the inherent hierarchical structures within each modality in hyperbolic space. To better integrate multimodal features in hyperbolic space, we introduce the ARCL module, which uses ranking-based contrastive learning to preserve the ordinal nature of survival time, along with the CUC module to fully explore the censored data. Extensive experiments demonstrate that our method outperforms state-of-the-art methods on five benchmark datasets. The source code is to be released.
MedKAN: An Advanced Kolmogorov-Arnold Network for Medical Image Classification
Feb 25, 2025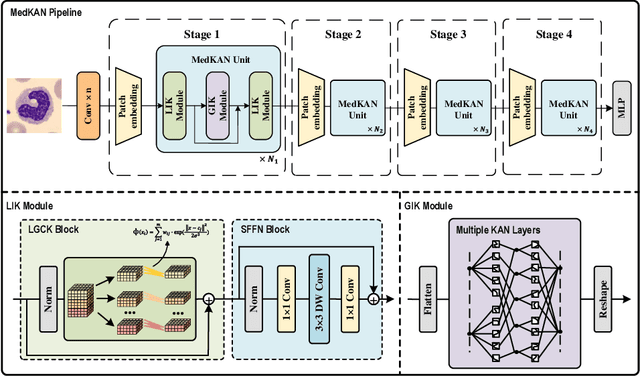
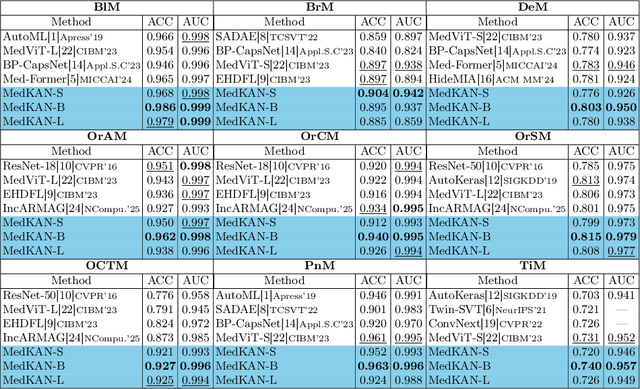
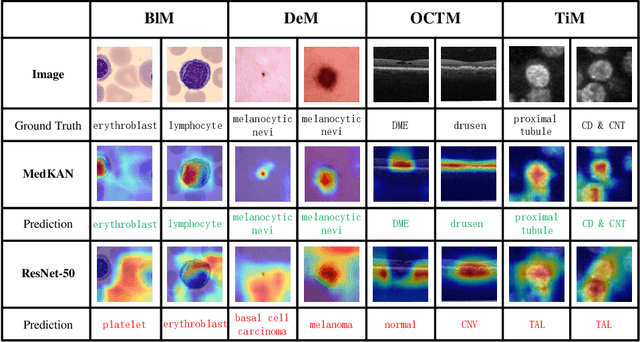
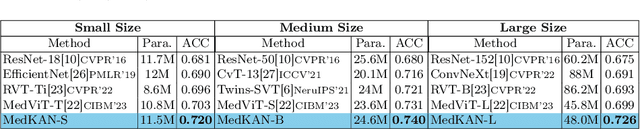
Abstract:Recent advancements in deep learning for image classification predominantly rely on convolutional neural networks (CNNs) or Transformer-based architectures. However, these models face notable challenges in medical imaging, particularly in capturing intricate texture details and contextual features. Kolmogorov-Arnold Networks (KANs) represent a novel class of architectures that enhance nonlinear transformation modeling, offering improved representation of complex features. In this work, we present MedKAN, a medical image classification framework built upon KAN and its convolutional extensions. MedKAN features two core modules: the Local Information KAN (LIK) module for fine-grained feature extraction and the Global Information KAN (GIK) module for global context integration. By combining these modules, MedKAN achieves robust feature modeling and fusion. To address diverse computational needs, we introduce three scalable variants--MedKAN-S, MedKAN-B, and MedKAN-L. Experimental results on nine public medical imaging datasets demonstrate that MedKAN achieves superior performance compared to CNN- and Transformer-based models, highlighting its effectiveness and generalizability in medical image analysis.
DAMPER: A Dual-Stage Medical Report Generation Framework with Coarse-Grained MeSH Alignment and Fine-Grained Hypergraph Matching
Dec 19, 2024



Abstract:Medical report generation is crucial for clinical diagnosis and patient management, summarizing diagnoses and recommendations based on medical imaging. However, existing work often overlook the clinical pipeline involved in report writing, where physicians typically conduct an initial quick review followed by a detailed examination. Moreover, current alignment methods may lead to misaligned relationships. To address these issues, we propose DAMPER, a dual-stage framework for medical report generation that mimics the clinical pipeline of report writing in two stages. In the first stage, a MeSH-Guided Coarse-Grained Alignment (MCG) stage that aligns chest X-ray (CXR) image features with medical subject headings (MeSH) features to generate a rough keyphrase representation of the overall impression. In the second stage, a Hypergraph-Enhanced Fine-Grained Alignment (HFG) stage that constructs hypergraphs for image patches and report annotations, modeling high-order relationships within each modality and performing hypergraph matching to capture semantic correlations between image regions and textual phrases. Finally,the coarse-grained visual features, generated MeSH representations, and visual hypergraph features are fed into a report decoder to produce the final medical report. Extensive experiments on public datasets demonstrate the effectiveness of DAMPER in generating comprehensive and accurate medical reports, outperforming state-of-the-art methods across various evaluation metrics.
Activation Space Selectable Kolmogorov-Arnold Networks
Aug 15, 2024



Abstract:The multilayer perceptron (MLP), a fundamental paradigm in current artificial intelligence, is widely applied in fields such as computer vision and natural language processing. However, the recently proposed Kolmogorov-Arnold Network (KAN), based on nonlinear additive connections, has been proven to achieve performance comparable to MLPs with significantly fewer parameters. Despite this potential, the use of a single activation function space results in reduced performance of KAN and related works across different tasks. To address this issue, we propose an activation space Selectable KAN (S-KAN). S-KAN employs an adaptive strategy to choose the possible activation mode for data at each feedforward KAN node. Our approach outperforms baseline methods in seven representative function fitting tasks and significantly surpasses MLP methods with the same level of parameters. Furthermore, we extend the structure of S-KAN and propose an activation space selectable Convolutional KAN (S-ConvKAN), which achieves leading results on four general image classification datasets. Our method mitigates the performance variability of the original KAN across different tasks and demonstrates through extensive experiments that feedforward KANs with selectable activations can achieve or even exceed the performance of MLP-based methods. This work contributes to the understanding of the data-centric design of new AI paradigms and provides a foundational reference for innovations in KAN-based network architectures.
GEM: Context-Aware Gaze EstiMation with Visual Search Behavior Matching for Chest Radiograph
Aug 10, 2024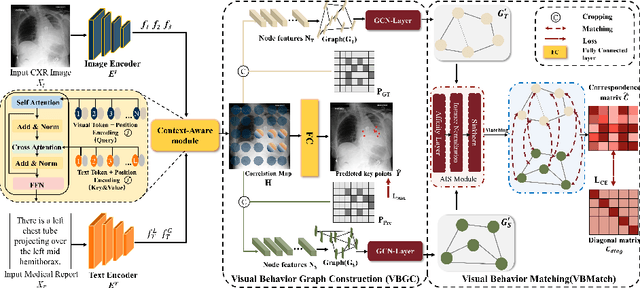
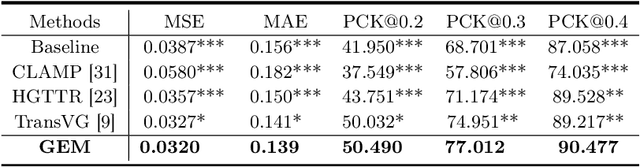
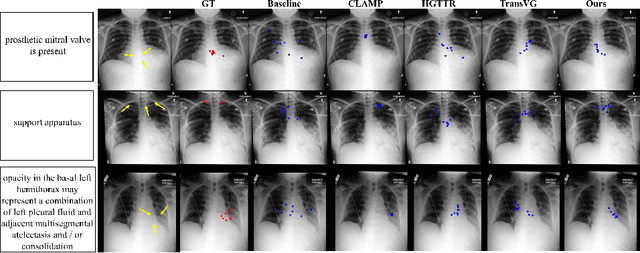
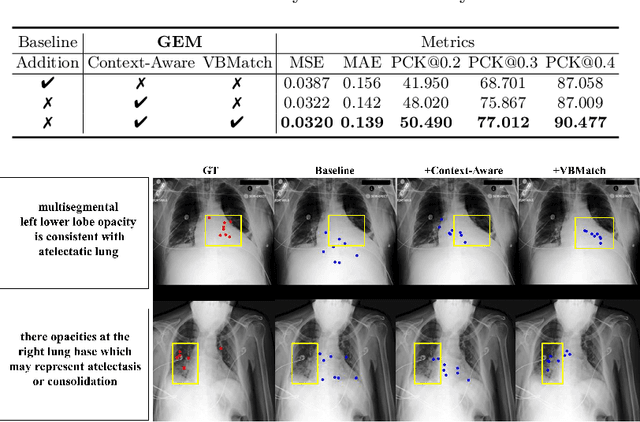
Abstract:Gaze estimation is pivotal in human scene comprehension tasks, particularly in medical diagnostic analysis. Eye-tracking technology facilitates the recording of physicians' ocular movements during image interpretation, thereby elucidating their visual attention patterns and information-processing strategies. In this paper, we initially define the context-aware gaze estimation problem in medical radiology report settings. To understand the attention allocation and cognitive behavior of radiologists during the medical image interpretation process, we propose a context-aware Gaze EstiMation (GEM) network that utilizes eye gaze data collected from radiologists to simulate their visual search behavior patterns throughout the image interpretation process. It consists of a context-awareness module, visual behavior graph construction, and visual behavior matching. Within the context-awareness module, we achieve intricate multimodal registration by establishing connections between medical reports and images. Subsequently, for a more accurate simulation of genuine visual search behavior patterns, we introduce a visual behavior graph structure, capturing such behavior through high-order relationships (edges) between gaze points (nodes). To maintain the authenticity of visual behavior, we devise a visual behavior-matching approach, adjusting the high-order relationships between them by matching the graph constructed from real and estimated gaze points. Extensive experiments on four publicly available datasets demonstrate the superiority of GEM over existing methods and its strong generalizability, which also provides a new direction for the effective utilization of diverse modalities in medical image interpretation and enhances the interpretability of models in the field of medical imaging. https://github.com/Tiger-SN/GEM
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge