Huayu Wang
Detector-in-the-Loop Tracking: Active Memory Rectification for Stable Glottic Opening Localization
Feb 22, 2026Abstract:Temporal stability in glottic opening localization remains challenging due to the complementary weaknesses of single-frame detectors and foundation-model trackers: the former lacks temporal context, while the latter suffers from memory drift. Specifically, in video laryngoscopy, rapid tissue deformation, occlusions, and visual ambiguities in emergency settings require a robust, temporally aware solution that can prevent progressive tracking errors. We propose Closed-Loop Memory Correction (CL-MC), a detector-in-the-loop framework that supervises Segment Anything Model 2(SAM2) through confidence-aligned state decisions and active memory rectification. High-confidence detections trigger semantic resets that overwrite corrupted tracker memory, effectively mitigating drift accumulation with a training-free foundation tracker in complex endoscopic scenes. On emergency intubation videos, CL-MC achieves state-of-the-art performance, significantly reducing drift and missing rate compared with the SAM2 variants and open loop based methods. Our results establish memory correction as a crucial component for reliable clinical video tracking. Our code will be available in https://github.com/huayuww/CL-MR.
Towards Globally Predictable k-Space Interpolation: A White-box Transformer Approach
Aug 06, 2025Abstract:Interpolating missing data in k-space is essential for accelerating imaging. However, existing methods, including convolutional neural network-based deep learning, primarily exploit local predictability while overlooking the inherent global dependencies in k-space. Recently, Transformers have demonstrated remarkable success in natural language processing and image analysis due to their ability to capture long-range dependencies. This inspires the use of Transformers for k-space interpolation to better exploit its global structure. However, their lack of interpretability raises concerns regarding the reliability of interpolated data. To address this limitation, we propose GPI-WT, a white-box Transformer framework based on Globally Predictable Interpolation (GPI) for k-space. Specifically, we formulate GPI from the perspective of annihilation as a novel k-space structured low-rank (SLR) model. The global annihilation filters in the SLR model are treated as learnable parameters, and the subgradients of the SLR model naturally induce a learnable attention mechanism. By unfolding the subgradient-based optimization algorithm of SLR into a cascaded network, we construct the first white-box Transformer specifically designed for accelerated MRI. Experimental results demonstrate that the proposed method significantly outperforms state-of-the-art approaches in k-space interpolation accuracy while providing superior interpretability.
TEMPURA: Temporal Event Masked Prediction and Understanding for Reasoning in Action
May 02, 2025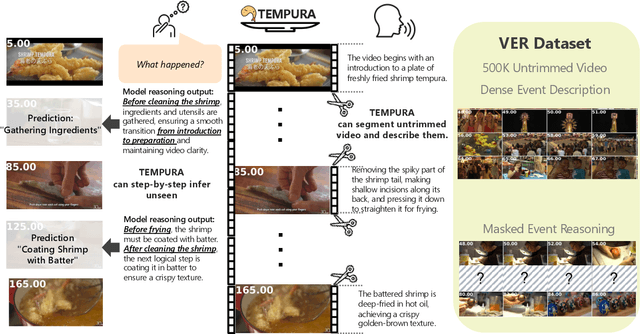
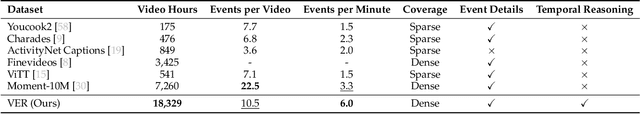
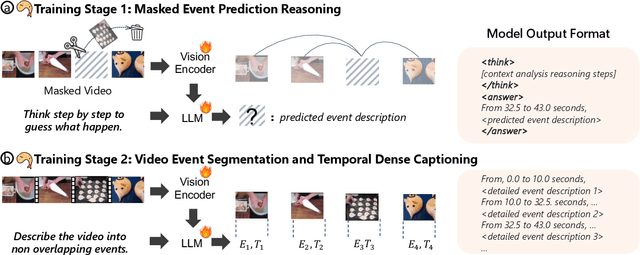
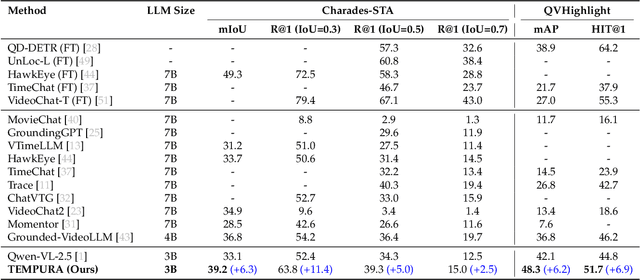
Abstract:Understanding causal event relationships and achieving fine-grained temporal grounding in videos remain challenging for vision-language models. Existing methods either compress video tokens to reduce temporal resolution, or treat videos as unsegmented streams, which obscures fine-grained event boundaries and limits the modeling of causal dependencies. We propose TEMPURA (Temporal Event Masked Prediction and Understanding for Reasoning in Action), a two-stage training framework that enhances video temporal understanding. TEMPURA first applies masked event prediction reasoning to reconstruct missing events and generate step-by-step causal explanations from dense event annotations, drawing inspiration from effective infilling techniques. TEMPURA then learns to perform video segmentation and dense captioning to decompose videos into non-overlapping events with detailed, timestamp-aligned descriptions. We train TEMPURA on VER, a large-scale dataset curated by us that comprises 1M training instances and 500K videos with temporally aligned event descriptions and structured reasoning steps. Experiments on temporal grounding and highlight detection benchmarks demonstrate that TEMPURA outperforms strong baseline models, confirming that integrating causal reasoning with fine-grained temporal segmentation leads to improved video understanding.
CrossFusion: A Multi-Scale Cross-Attention Convolutional Fusion Model for Cancer Survival Prediction
Mar 03, 2025Abstract:Cancer survival prediction from whole slide images (WSIs) is a challenging task in computational pathology due to the large size, irregular shape, and high granularity of the WSIs. These characteristics make it difficult to capture the full spectrum of patterns, from subtle cellular abnormalities to complex tissue interactions, which are crucial for accurate prognosis. To address this, we propose CrossFusion, a novel multi-scale feature integration framework that extracts and fuses information from patches across different magnification levels. By effectively modeling both scale-specific patterns and their interactions, CrossFusion generates a rich feature set that enhances survival prediction accuracy. We validate our approach across six cancer types from public datasets, demonstrating significant improvements over existing state-of-the-art methods. Moreover, when coupled with domain-specific feature extraction backbones, our method shows further gains in prognostic performance compared to general-purpose backbones. The source code is available at: https://github.com/RustinS/CrossFusion
Joint PET-MRI Reconstruction with Diffusion Stochastic Differential Model
Aug 07, 2024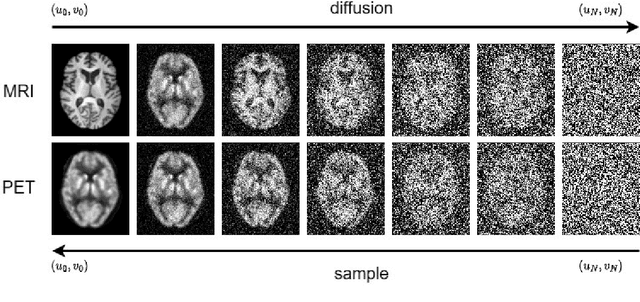
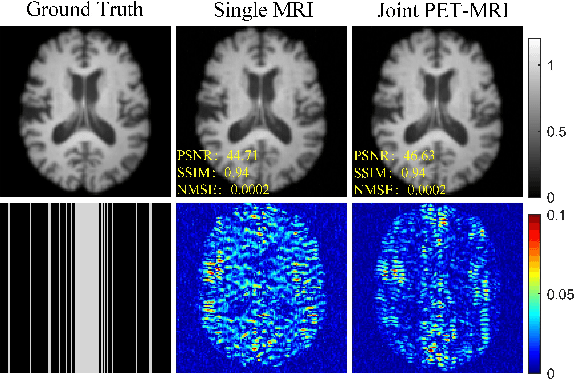
Abstract:PET suffers from a low signal-to-noise ratio. Meanwhile, the k-space data acquisition process in MRI is time-consuming by PET-MRI systems. We aim to accelerate MRI and improve PET image quality. This paper proposed a novel joint reconstruction model by diffusion stochastic differential equations based on learning the joint probability distribution of PET and MRI. Compare the results underscore the qualitative and quantitative improvements our model brings to PET and MRI reconstruction, surpassing the current state-of-the-art methodologies. Joint PET-MRI reconstruction is a challenge in the PET-MRI system. This studies focused on the relationship extends beyond edges. In this study, PET is generated from MRI by learning joint probability distribution as the relationship.
* Accepted as ISMRM 2024 Digital poster 6575. 04-09 May 2024 Singapore
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
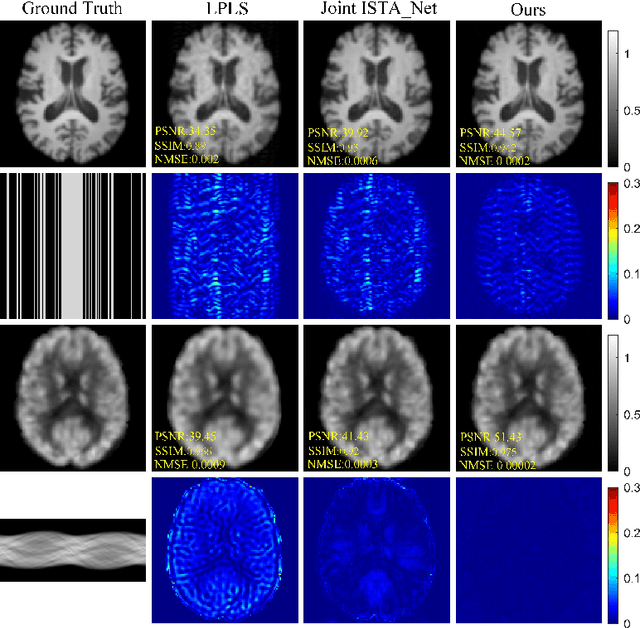
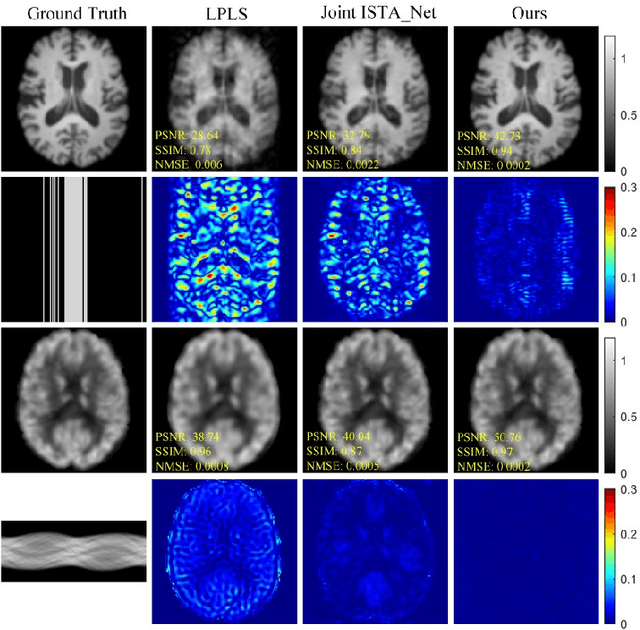
Joint Diffusion: Mutual Consistency-Driven Diffusion Model for PET-MRI Co-Reconstruction
Nov 24, 2023Abstract:Positron Emission Tomography and Magnetic Resonance Imaging (PET-MRI) systems can obtain functional and anatomical scans. PET suffers from a low signal-to-noise ratio. Meanwhile, the k-space data acquisition process in MRI is time-consuming. The study aims to accelerate MRI and enhance PET image quality. Conventional approaches involve the separate reconstruction of each modality within PET-MRI systems. However, there exists complementary information among multi-modal images. The complementary information can contribute to image reconstruction. In this study, we propose a novel PET-MRI joint reconstruction model employing a mutual consistency-driven diffusion mode, namely MC-Diffusion. MC-Diffusion learns the joint probability distribution of PET and MRI for utilizing complementary information. We conducted a series of contrast experiments about LPLS, Joint ISAT-net and MC-Diffusion by the ADNI dataset. The results underscore the qualitative and quantitative improvements achieved by MC-Diffusion, surpassing the state-of-the-art method.
Matrix Completion-Informed Deep Unfolded Equilibrium Models for Self-Supervised k-Space Interpolation in MRI
Sep 24, 2023



Abstract:Recently, regularization model-driven deep learning (DL) has gained significant attention due to its ability to leverage the potent representational capabilities of DL while retaining the theoretical guarantees of regularization models. However, most of these methods are tailored for supervised learning scenarios that necessitate fully sampled labels, which can pose challenges in practical MRI applications. To tackle this challenge, we propose a self-supervised DL approach for accelerated MRI that is theoretically guaranteed and does not rely on fully sampled labels. Specifically, we achieve neural network structure regularization by exploiting the inherent structural low-rankness of the $k$-space data. Simultaneously, we constrain the network structure to resemble a nonexpansive mapping, ensuring the network's convergence to a fixed point. Thanks to this well-defined network structure, this fixed point can completely reconstruct the missing $k$-space data based on matrix completion theory, even in situations where full-sampled labels are unavailable. Experiments validate the effectiveness of our proposed method and demonstrate its superiority over existing self-supervised approaches and traditional regularization methods, achieving performance comparable to that of supervised learning methods in certain scenarios.
Convex Latent-Optimized Adversarial Regularizers for Imaging Inverse Problems
Sep 17, 2023



Abstract:Recently, data-driven techniques have demonstrated remarkable effectiveness in addressing challenges related to MR imaging inverse problems. However, these methods still exhibit certain limitations in terms of interpretability and robustness. In response, we introduce Convex Latent-Optimized Adversarial Regularizers (CLEAR), a novel and interpretable data-driven paradigm. CLEAR represents a fusion of deep learning (DL) and variational regularization. Specifically, we employ a latent optimization technique to adversarially train an input convex neural network, and its set of minima can fully represent the real data manifold. We utilize it as a convex regularizer to formulate a CLEAR-informed variational regularization model that guides the solution of the imaging inverse problem on the real data manifold. Leveraging its inherent convexity, we have established the convergence of the projected subgradient descent algorithm for the CLEAR-informed regularization model. This convergence guarantees the attainment of a unique solution to the imaging inverse problem, subject to certain assumptions. Furthermore, we have demonstrated the robustness of our CLEAR-informed model, explicitly showcasing its capacity to achieve stable reconstruction even in the presence of measurement interference. Finally, we illustrate the superiority of our approach using MRI reconstruction as an example. Our method consistently outperforms conventional data-driven techniques and traditional regularization approaches, excelling in both reconstruction quality and robustness.
A Survey on Label-efficient Deep Segmentation: Bridging the Gap between Weak Supervision and Dense Prediction
Jul 04, 2022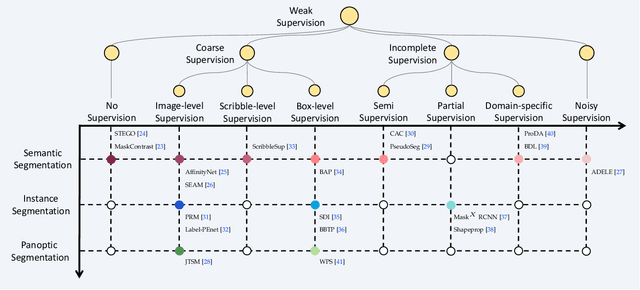
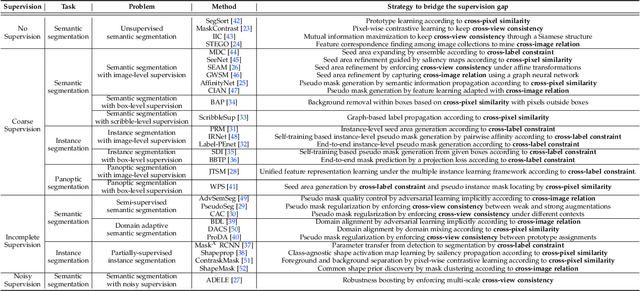

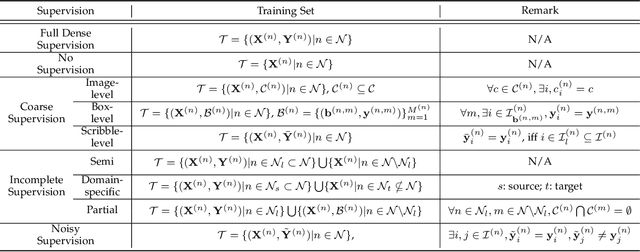
Abstract:The rapid development of deep learning has made a great progress in segmentation, one of the fundamental tasks of computer vision. However, the current segmentation algorithms mostly rely on the availability of pixel-level annotations, which are often expensive, tedious, and laborious. To alleviate this burden, the past years have witnessed an increasing attention in building label-efficient, deep-learning-based segmentation algorithms. This paper offers a comprehensive review on label-efficient segmentation methods. To this end, we first develop a taxonomy to organize these methods according to the supervision provided by different types of weak labels (including no supervision, coarse supervision, incomplete supervision and noisy supervision) and supplemented by the types of segmentation problems (including semantic segmentation, instance segmentation and panoptic segmentation). Next, we summarize the existing label-efficient segmentation methods from a unified perspective that discusses an important question: how to bridge the gap between weak supervision and dense prediction -- the current methods are mostly based on heuristic priors, such as cross-pixel similarity, cross-label constraint, cross-view consistency, cross-image relation, etc. Finally, we share our opinions about the future research directions for label-efficient deep segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge