Linda Shapiro
Detector-in-the-Loop Tracking: Active Memory Rectification for Stable Glottic Opening Localization
Feb 22, 2026Abstract:Temporal stability in glottic opening localization remains challenging due to the complementary weaknesses of single-frame detectors and foundation-model trackers: the former lacks temporal context, while the latter suffers from memory drift. Specifically, in video laryngoscopy, rapid tissue deformation, occlusions, and visual ambiguities in emergency settings require a robust, temporally aware solution that can prevent progressive tracking errors. We propose Closed-Loop Memory Correction (CL-MC), a detector-in-the-loop framework that supervises Segment Anything Model 2(SAM2) through confidence-aligned state decisions and active memory rectification. High-confidence detections trigger semantic resets that overwrite corrupted tracker memory, effectively mitigating drift accumulation with a training-free foundation tracker in complex endoscopic scenes. On emergency intubation videos, CL-MC achieves state-of-the-art performance, significantly reducing drift and missing rate compared with the SAM2 variants and open loop based methods. Our results establish memory correction as a crucial component for reliable clinical video tracking. Our code will be available in https://github.com/huayuww/CL-MR.
Unified and Semantically Grounded Domain Adaptation for Medical Image Segmentation
Aug 12, 2025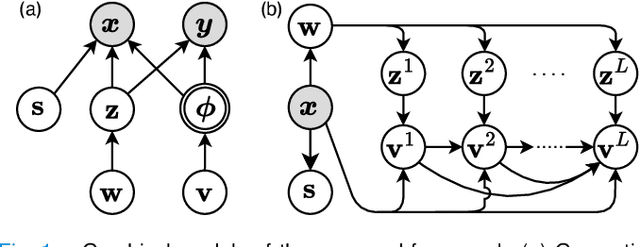
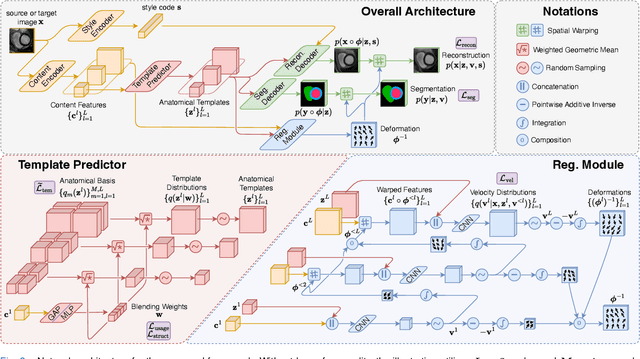
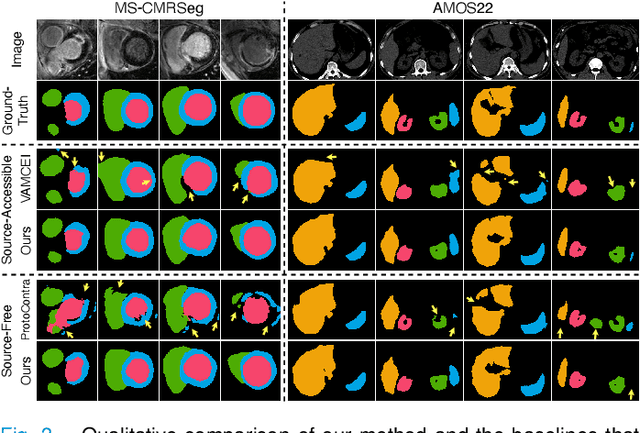
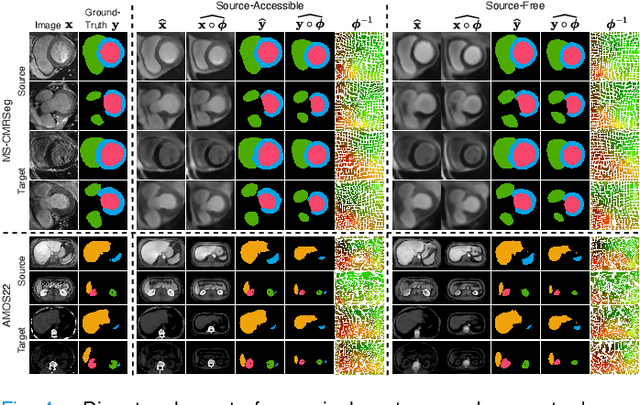
Abstract:Most prior unsupervised domain adaptation approaches for medical image segmentation are narrowly tailored to either the source-accessible setting, where adaptation is guided by source-target alignment, or the source-free setting, which typically resorts to implicit supervision mechanisms such as pseudo-labeling and model distillation. This substantial divergence in methodological designs between the two settings reveals an inherent flaw: the lack of an explicit, structured construction of anatomical knowledge that naturally generalizes across domains and settings. To bridge this longstanding divide, we introduce a unified, semantically grounded framework that supports both source-accessible and source-free adaptation. Fundamentally distinct from all prior works, our framework's adaptability emerges naturally as a direct consequence of the model architecture, without the need for any handcrafted adaptation strategies. Specifically, our model learns a domain-agnostic probabilistic manifold as a global space of anatomical regularities, mirroring how humans establish visual understanding. Thus, the structural content in each image can be interpreted as a canonical anatomy retrieved from the manifold and a spatial transformation capturing individual-specific geometry. This disentangled, interpretable formulation enables semantically meaningful prediction with intrinsic adaptability. Extensive experiments on challenging cardiac and abdominal datasets show that our framework achieves state-of-the-art results in both settings, with source-free performance closely approaching its source-accessible counterpart, a level of consistency rarely observed in prior works. Beyond quantitative improvement, we demonstrate strong interpretability of the proposed framework via manifold traversal for smooth shape manipulation.
BADGR: Bundle Adjustment Diffusion Conditioned by GRadients for Wide-Baseline Floor Plan Reconstruction
Mar 25, 2025Abstract:Reconstructing precise camera poses and floor plan layouts from wide-baseline RGB panoramas is a difficult and unsolved problem. We introduce BADGR, a novel diffusion model that jointly performs reconstruction and bundle adjustment (BA) to refine poses and layouts from a coarse state, using 1D floor boundary predictions from dozens of images of varying input densities. Unlike a guided diffusion model, BADGR is conditioned on dense per-entity outputs from a single-step Levenberg Marquardt (LM) optimizer and is trained to predict camera and wall positions while minimizing reprojection errors for view-consistency. The objective of layout generation from denoising diffusion process complements BA optimization by providing additional learned layout-structural constraints on top of the co-visible features across images. These constraints help BADGR to make plausible guesses on spatial relations which help constrain pose graph, such as wall adjacency, collinearity, and learn to mitigate errors from dense boundary observations with global contexts. BADGR trains exclusively on 2D floor plans, simplifying data acquisition, enabling robust augmentation, and supporting variety of input densities. Our experiments and analysis validate our method, which significantly outperforms the state-of-the-art pose and floor plan layout reconstruction with different input densities.
CrossFusion: A Multi-Scale Cross-Attention Convolutional Fusion Model for Cancer Survival Prediction
Mar 03, 2025Abstract:Cancer survival prediction from whole slide images (WSIs) is a challenging task in computational pathology due to the large size, irregular shape, and high granularity of the WSIs. These characteristics make it difficult to capture the full spectrum of patterns, from subtle cellular abnormalities to complex tissue interactions, which are crucial for accurate prognosis. To address this, we propose CrossFusion, a novel multi-scale feature integration framework that extracts and fuses information from patches across different magnification levels. By effectively modeling both scale-specific patterns and their interactions, CrossFusion generates a rich feature set that enhances survival prediction accuracy. We validate our approach across six cancer types from public datasets, demonstrating significant improvements over existing state-of-the-art methods. Moreover, when coupled with domain-specific feature extraction backbones, our method shows further gains in prognostic performance compared to general-purpose backbones. The source code is available at: https://github.com/RustinS/CrossFusion
PathFinder: A Multi-Modal Multi-Agent System for Medical Diagnostic Decision-Making Applied to Histopathology
Feb 13, 2025



Abstract:Diagnosing diseases through histopathology whole slide images (WSIs) is fundamental in modern pathology but is challenged by the gigapixel scale and complexity of WSIs. Trained histopathologists overcome this challenge by navigating the WSI, looking for relevant patches, taking notes, and compiling them to produce a final holistic diagnostic. Traditional AI approaches, such as multiple instance learning and transformer-based models, fail short of such a holistic, iterative, multi-scale diagnostic procedure, limiting their adoption in the real-world. We introduce PathFinder, a multi-modal, multi-agent framework that emulates the decision-making process of expert pathologists. PathFinder integrates four AI agents, the Triage Agent, Navigation Agent, Description Agent, and Diagnosis Agent, that collaboratively navigate WSIs, gather evidence, and provide comprehensive diagnoses with natural language explanations. The Triage Agent classifies the WSI as benign or risky; if risky, the Navigation and Description Agents iteratively focus on significant regions, generating importance maps and descriptive insights of sampled patches. Finally, the Diagnosis Agent synthesizes the findings to determine the patient's diagnostic classification. Our Experiments show that PathFinder outperforms state-of-the-art methods in skin melanoma diagnosis by 8% while offering inherent explainability through natural language descriptions of diagnostically relevant patches. Qualitative analysis by pathologists shows that the Description Agent's outputs are of high quality and comparable to GPT-4o. PathFinder is also the first AI-based system to surpass the average performance of pathologists in this challenging melanoma classification task by 9%, setting a new record for efficient, accurate, and interpretable AI-assisted diagnostics in pathology. Data, code and models available at https://pathfinder-dx.github.io/
MedicalNarratives: Connecting Medical Vision and Language with Localized Narratives
Jan 07, 2025



Abstract:We propose MedicalNarratives, a dataset curated from medical pedagogical videos similar in nature to data collected in Think-Aloud studies and inspired by Localized Narratives, which collects grounded image-text data by curating instructors' speech and mouse cursor movements synchronized in time. MedicalNarratives enables pretraining of both semantic and dense objectives, alleviating the need to train medical semantic and dense tasks disparately due to the lack of reasonably sized datasets. Our dataset contains 4.7M image-text pairs from videos and articles, with 1M samples containing dense annotations in the form of traces and bounding boxes. To evaluate the utility of MedicalNarratives, we train GenMedClip based on the CLIP architecture using our dataset spanning 12 medical domains and demonstrate that it outperforms previous state-of-the-art models on a newly constructed medical imaging benchmark that comprehensively evaluates performance across all modalities. Data, demo, code and models available at https://medical-narratives.github.io
Generating Seamless Virtual Immunohistochemical Whole Slide Images with Content and Color Consistency
Oct 01, 2024



Abstract:Immunohistochemical (IHC) stains play a vital role in a pathologist's analysis of medical images, providing crucial diagnostic information for various diseases. Virtual staining from hematoxylin and eosin (H&E)-stained whole slide images (WSIs) allows the automatic production of other useful IHC stains without the expensive physical staining process. However, current virtual WSI generation methods based on tile-wise processing often suffer from inconsistencies in content, texture, and color at tile boundaries. These inconsistencies lead to artifacts that compromise image quality and potentially hinder accurate clinical assessment and diagnoses. To address this limitation, we propose a novel consistent WSI synthesis network, CC-WSI-Net, that extends GAN models to produce seamless synthetic whole slide images. Our CC-WSI-Net integrates a content- and color-consistency supervisor, ensuring consistency across tiles and facilitating the generation of seamless synthetic WSIs while ensuring Sox10 immunohistochemistry accuracy in melanocyte detection. We validate our method through extensive image-quality analyses, objective detection assessments, and a subjective survey with pathologists. By generating high-quality synthetic WSIs, our method opens doors for advanced virtual staining techniques with broader applications in research and clinical care.
Bayesian Intrinsic Groupwise Image Registration: Unsupervised Disentanglement of Anatomy and Geometry
Jan 04, 2024



Abstract:This article presents a general Bayesian learning framework for multi-modal groupwise registration on medical images. The method builds on probabilistic modelling of the image generative process, where the underlying common anatomy and geometric variations of the observed images are explicitly disentangled as latent variables. Thus, groupwise registration is achieved through the solution to Bayesian inference. We propose a novel hierarchical variational auto-encoding architecture to realize the inference procedure of the latent variables, where the registration parameters can be calculated in a mathematically interpretable fashion. Remarkably, this new paradigm can learn groupwise registration in an unsupervised closed-loop self-reconstruction process, sparing the burden of designing complex intensity-based similarity measures. The computationally efficient disentangled architecture is also inherently scalable and flexible, allowing for groupwise registration on large-scale image groups with variable sizes. Furthermore, the inferred structural representations from disentanglement learning are capable of capturing the latent anatomy of the observations with visual semantics. Extensive experiments were conducted to validate the proposed framework, including four datasets from cardiac, brain and abdominal medical images. The results have demonstrated the superiority of our method over conventional similarity-based approaches in terms of accuracy, efficiency, scalability and interpretability.
Quilt-LLaVA: Visual Instruction Tuning by Extracting Localized Narratives from Open-Source Histopathology Videos
Dec 07, 2023Abstract:The gigapixel scale of whole slide images (WSIs) poses a challenge for histopathology multi-modal chatbots, requiring a global WSI analysis for diagnosis, compounding evidence from different WSI patches. Current visual instruction datasets, generated through large language models, focus on creating question/answer pairs for individual image patches, which may lack diagnostic capacity on their own in histopathology, further complicated by the absence of spatial grounding in histopathology image captions. To bridge this gap, we introduce Quilt-Instruct, a large-scale dataset of 107,131 histopathology-specific instruction question/answer pairs, that is collected by leveraging educational histopathology videos from YouTube, which provides spatial localization of captions by automatically extracting narrators' cursor movements. In addition, we provide contextual reasoning by extracting diagnosis and supporting facts from the entire video content to guide the extrapolative reasoning of GPT-4. Using Quilt-Instruct, we train Quilt-LLaVA, which can reason beyond the given single image patch, enabling diagnostic reasoning and the capability of spatial awareness. To evaluate Quilt-LLaVA, we propose a comprehensive evaluation dataset created from 985 images and 1283 human-generated question-answers. We also thoroughly evaluate Quilt-LLaVA using public histopathology datasets, where Quilt-LLaVA significantly outperforms SOTA by over 10% on relative GPT-4 score and 4% and 9% on open and closed set VQA. Our code, data, and model are publicly available at quilt-llava.github.io.
Quilt-1M: One Million Image-Text Pairs for Histopathology
Jun 22, 2023Abstract:Recent accelerations in multi-modal applications have been made possible with the plethora of image and text data available online. However, the scarcity of analogous data in the medical field, specifically in histopathology, has halted comparable progress. To enable similar representation learning for histopathology, we turn to YouTube, an untapped resource of videos, offering $1,087$ hours of valuable educational histopathology videos from expert clinicians. From YouTube, we curate Quilt: a large-scale vision-language dataset consisting of $768,826$ image and text pairs. Quilt was automatically curated using a mixture of models, including large language models, handcrafted algorithms, human knowledge databases, and automatic speech recognition. In comparison, the most comprehensive datasets curated for histopathology amass only around $200$K samples. We combine Quilt with datasets from other sources, including Twitter, research papers, and the internet in general, to create an even larger dataset: Quilt-1M, with $1$M paired image-text samples, marking it as the largest vision-language histopathology dataset to date. We demonstrate the value of Quilt-1M by fine-tuning a pre-trained CLIP model. Our model outperforms state-of-the-art models on both zero-shot and linear probing tasks for classifying new histopathology images across $13$ diverse patch-level datasets of $8$ different sub-pathologies and cross-modal retrieval tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge