Wisdom Oluchi Ikezogwo
Quilt-1M: One Million Image-Text Pairs for Histopathology
Jun 22, 2023Abstract:Recent accelerations in multi-modal applications have been made possible with the plethora of image and text data available online. However, the scarcity of analogous data in the medical field, specifically in histopathology, has halted comparable progress. To enable similar representation learning for histopathology, we turn to YouTube, an untapped resource of videos, offering $1,087$ hours of valuable educational histopathology videos from expert clinicians. From YouTube, we curate Quilt: a large-scale vision-language dataset consisting of $768,826$ image and text pairs. Quilt was automatically curated using a mixture of models, including large language models, handcrafted algorithms, human knowledge databases, and automatic speech recognition. In comparison, the most comprehensive datasets curated for histopathology amass only around $200$K samples. We combine Quilt with datasets from other sources, including Twitter, research papers, and the internet in general, to create an even larger dataset: Quilt-1M, with $1$M paired image-text samples, marking it as the largest vision-language histopathology dataset to date. We demonstrate the value of Quilt-1M by fine-tuning a pre-trained CLIP model. Our model outperforms state-of-the-art models on both zero-shot and linear probing tasks for classifying new histopathology images across $13$ diverse patch-level datasets of $8$ different sub-pathologies and cross-modal retrieval tasks.
Multi-modal Masked Autoencoders Learn Compositional Histopathological Representations
Sep 04, 2022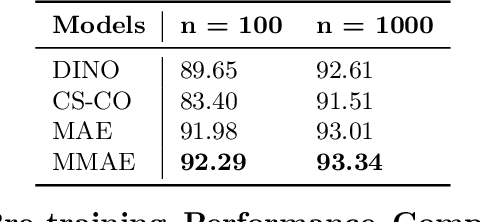
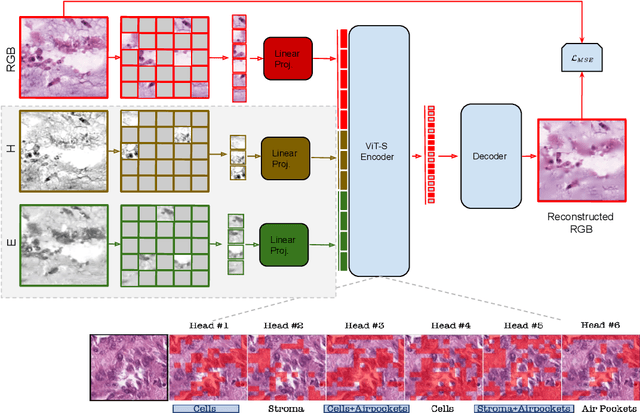
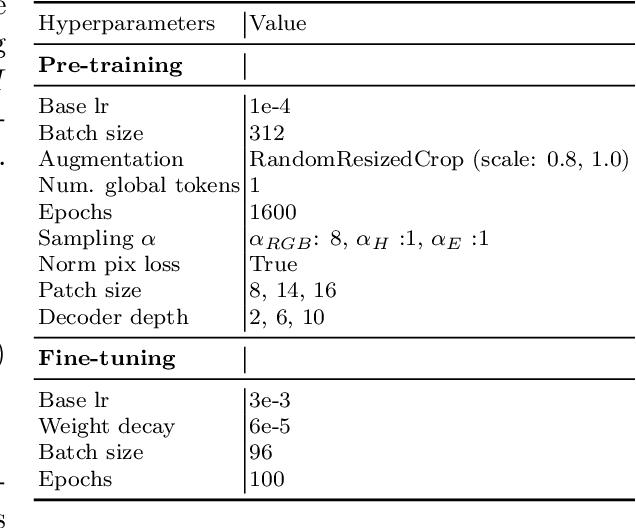
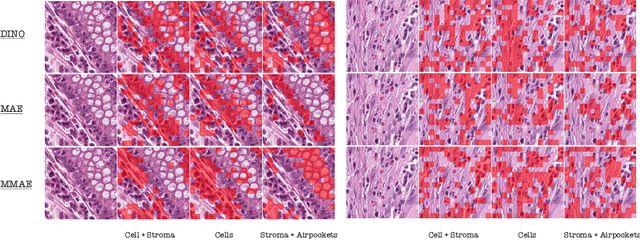
Abstract:Self-supervised learning (SSL) enables learning useful inductive biases through utilizing pretext tasks that require no labels. The unlabeled nature of SSL makes it especially important for whole slide histopathological images (WSIs), where patch-level human annotation is difficult. Masked Autoencoders (MAE) is a recent SSL method suitable for digital pathology as it does not require negative sampling and requires little to no data augmentations. However, the domain shift between natural images and digital pathology images requires further research in designing MAE for patch-level WSIs. In this paper, we investigate several design choices for MAE in histopathology. Furthermore, we introduce a multi-modal MAE (MMAE) that leverages the specific compositionality of Hematoxylin & Eosin (H&E) stained WSIs. We performed our experiments on the public patch-level dataset NCT-CRC-HE-100K. The results show that the MMAE architecture outperforms supervised baselines and other state-of-the-art SSL techniques for an eight-class tissue phenotyping task, utilizing only 100 labeled samples for fine-tuning. Our code is available at https://github.com/wisdomikezogwo/MMAE_Pathology
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge