Xiaoliang Zhang
The UTE and ZTE Sequences at Ultra-High Magnetic Field Strengths: A Survey
Oct 07, 2022
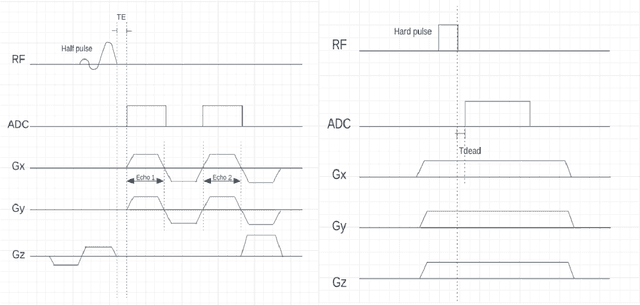
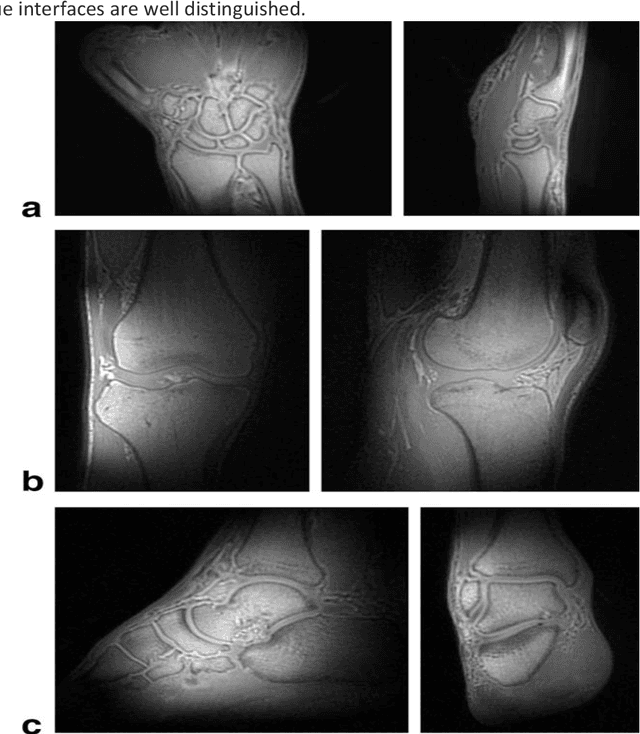

Abstract:UTE (Ultrashort Echo Time) and ZTE (Zero Echo Time) sequences have been developed to detect short T2 relaxation signals coming from regions that are unable to be detected by conventional MRI methods. Due to the high dipole-dipole interactions in solid and semi-solid tissues, the echo time generated is simply not enough to produce a signal using conventional imaging method, often leading to void signal coming from the discussed areas. By the application of these techniques, solid and semi-solid areas can be imaged which can have a profound impact in clinical imaging. High and Ultra-high field strength (UHF) provides a vital advantage in providing better sensitivity and specificity of MR imaging. When coupled with the UTE and ZTE sequences, the image can recover void signals as well as a much-improved signal quality. To further this strategy, secondary data from various research tools was obtained to further validate the research while addressing the drawbacks to this approach. It was found that UTE and ZTE sequences coupled with some techniques such as qualitative imaging and new trajectories are very crucial for accurate image depiction of the areas of the musculoskeletal system, neural system, lung imaging and dental imaging.
Unsupervised Deep Unrolled Reconstruction Using Regularization by Denoising
May 07, 2022



Abstract:Deep learning methods have been successfully used in various computer vision tasks. Inspired by that success, deep learning has been explored in magnetic resonance imaging (MRI) reconstruction. In particular, integrating deep learning and model-based optimization methods has shown considerable advantages. However, a large amount of labeled training data is typically needed for high reconstruction quality, which is challenging for some MRI applications. In this paper, we propose a novel reconstruction method, named DURED-Net, that enables interpretable unsupervised learning for MR image reconstruction by combining an unsupervised denoising network and a plug-and-play method. We aim to boost the reconstruction performance of unsupervised learning by adding an explicit prior that utilizes imaging physics. Specifically, the leverage of a denoising network for MRI reconstruction is achieved using Regularization by Denoising (RED). Experiment results demonstrate that the proposed method requires a reduced amount of training data to achieve high reconstruction quality.
Hairpin RF Resonators for Transceiver Arrays with High Inter-channel Isolation and B1 Efficiency at Ultrahigh Field 7T MR Imaging
Jan 21, 2022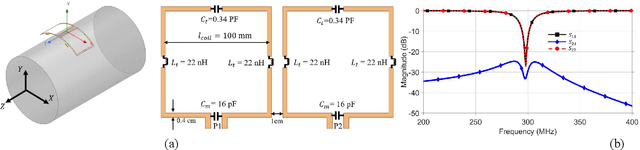
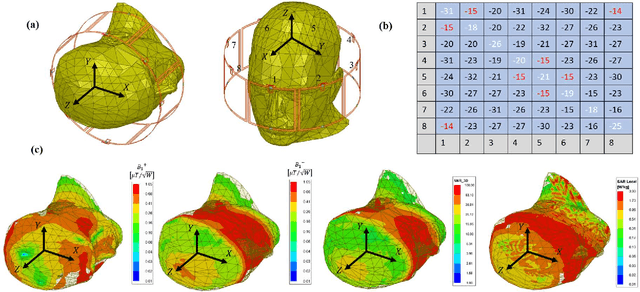
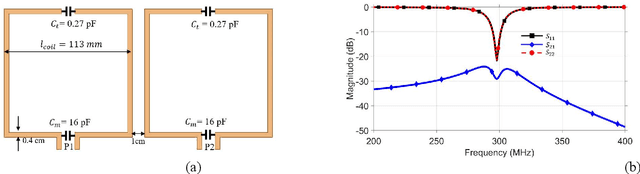
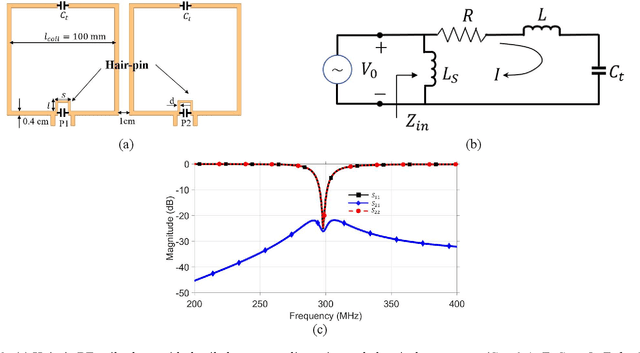
Abstract:Sufficient electromagnetic decoupling among resonant elements in RF array coils is required to maintain the integrity of the magnetic flux density map from individual channel at ultra-high field magnetic resonance imaging. A close-fitting or high-density transceiver array for augmented performance in parallel imaging and imaging quality, leads to strong coupling between elements of the array making the decoupling a very challenging task at ultra-high fields. High impedance RF coils have demonstrated to be a prominent design method to circumvent these coupling issues. However, inherent characteristics of these coils have ramification on the B1 field efficiency and SNR or the complexity of the design. In this work, we propose a hairpin RF coil, a simple design based on high impedance technique that provides excellent decoupling performance and superior RF magnetic field efficiency compared to the current state high impedance coils. In order to validate the feasibility of the proposed hairpin RF coils, systematical studies on decoupling performance, field distribution, and SNR are performed, and the results are compared with those obtained from one high impedance RF coil namely self-decoupled RF coil. To investigate the proposed hairpin RF coil design, a 7T 8-channel head coil array using hairpin resonators is built and evaluated. The MR imaging results in a cylindrical phantom obtained from the 8-channel array demonstrated a 13 % increase in SNR field intensity of the hairpin design compared to the self-decoupled coils at 7T under the same circumstance. Furthermore, the characteristics of the hairpin RF coils are evaluated using a more realistic human head voxel model in ANSYS HFSS.
Decoupled Self Attention for Accurate One Stage Object Detection
Dec 15, 2020
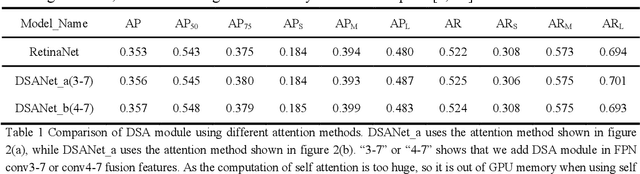
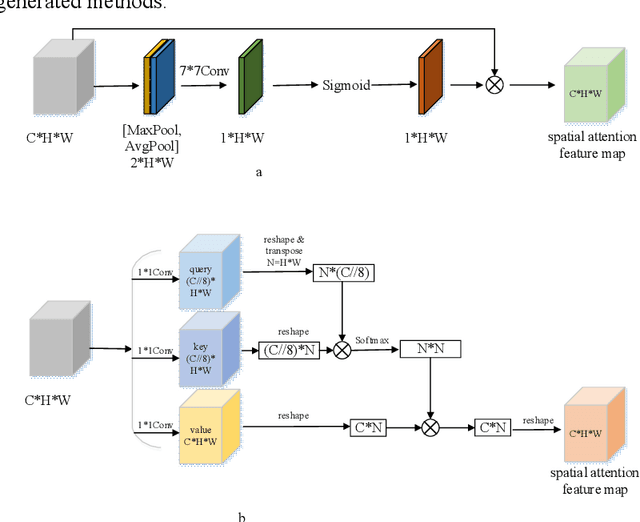

Abstract:As the scale of object detection dataset is smaller than that of image recognition dataset ImageNet, transfer learning has become a basic training method for deep learning object detection models, which will pretrain the backbone network of object detection model on ImageNet dataset to extract features for classification and localization subtasks. However, the classification task focuses on the salient region features of object, while the location task focuses on the edge features of object, so there is certain deviation between the features extracted by pretrained backbone network and the features used for localization task. In order to solve this problem, a decoupled self attention(DSA) module is proposed for one stage object detection models in this paper. DSA includes two decoupled self-attention branches, so it can extract appropriate features for different tasks. It is located between FPN and head networks of subtasks, so it is used to extract global features based on FPN fused features for different tasks independently. Although the network of DSA module is simple, but it can effectively improve the performance of object detection, also it can be easily embedded in many detection models. Our experiments are based on the representative one-stage detection model RetinaNet. In COCO dataset, when ResNet50 and ResNet101 are used as backbone networks, the detection performances can be increased by 0.4% AP and 0.5% AP respectively. When DSA module and object confidence task are applied in RetinaNet together, the detection performances based on ResNet50 and ResNet101 can be increased by 1.0% AP and 1.4% AP respectively. The experiment results show the effectiveness of DSA module. Code is at: https://github.com/chenzuge1/DSANet.git.
Deep Learning for Highly Accelerated Diffusion Tensor Imaging
Feb 03, 2020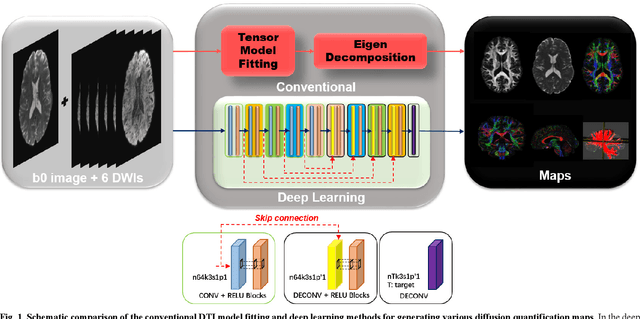
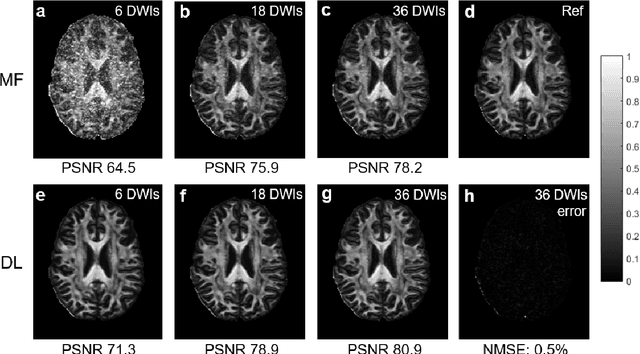
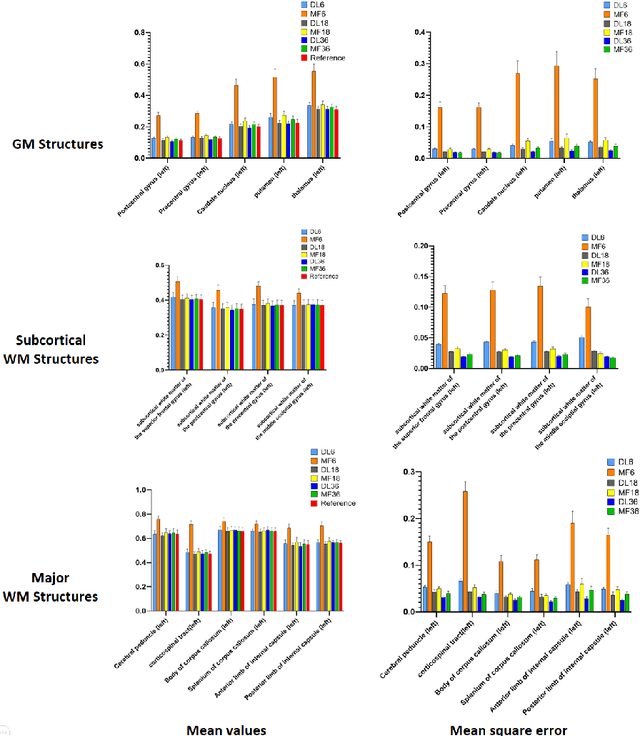

Abstract:Diffusion tensor imaging (DTI) is widely used to examine the human brain white matter structures, including their microarchitecture integrity and spatial fiber tract trajectories, with clinical applications in several neurological disorders and neurosurgical guidance. However, a major factor that prevents DTI from being incorporated in clinical routines is its long scan time due to the acquisition of a large number (typically 30 or more) of diffusion-weighted images (DWIs) required for reliable tensor estimation. Here, a deep learning-based technique is developed to obtain diffusion tensor images with only six DWIs, resulting in a significant reduction in imaging time. The method uses deep convolutional neural networks to learn the highly nonlinear relationship between DWIs and several tensor-derived maps, bypassing the conventional tensor fitting procedure, which is well known to be highly susceptible to noises in DWIs. The performance of the method was evaluated using DWI datasets from the Human Connectome Project and patients with ischemic stroke. Our results demonstrate that the proposed technique is able to generate quantitative maps of good quality fractional anisotropy (FA) and mean diffusivity (MD), as well as the fiber tractography from as few as six DWIs. The proposed method achieves a quantification error of less than 5% in all regions of interest of the brain, which is the rate of in vivo reproducibility of diffusion tensor imaging. Tractography reconstruction is also comparable to the ground truth obtained from 90 DWIs. In addition, we also demonstrate that the neural network trained on healthy volunteers can be directly applied/tested on stroke patients' DWIs data without compromising the lesion detectability. Such a significant reduction in scan time will allow inclusion of DTI into clinical routine for many potential applications.
A Random Sample Partition Data Model for Big Data Analysis
Jan 20, 2018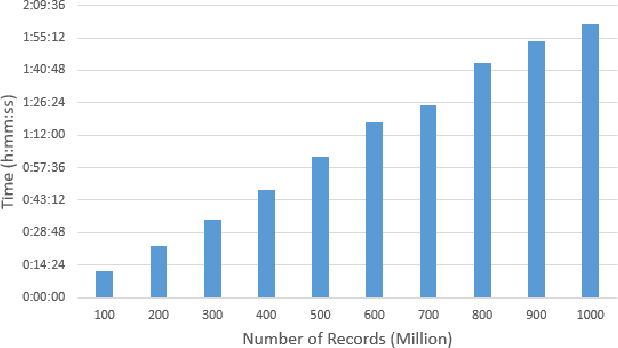
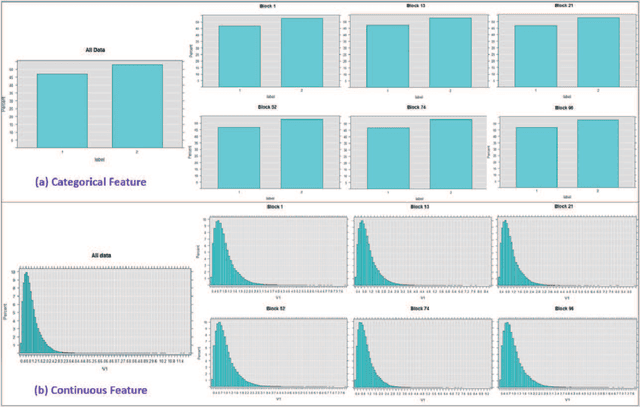

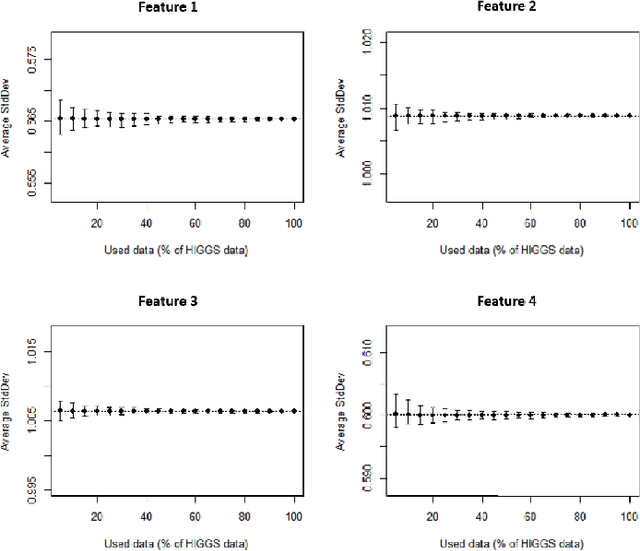
Abstract:Big data sets must be carefully partitioned into statistically similar data subsets that can be used as representative samples for big data analysis tasks. In this paper, we propose the random sample partition (RSP) data model to represent a big data set as a set of non-overlapping data subsets, called RSP data blocks, where each RSP data block has a probability distribution similar to the whole big data set. Under this data model, efficient block level sampling is used to randomly select RSP data blocks, replacing expensive record level sampling to select sample data from a big distributed data set on a computing cluster. We show how RSP data blocks can be employed to estimate statistics of a big data set and build models which are equivalent to those built from the whole big data set. In this approach, analysis of a big data set becomes analysis of few RSP data blocks which have been generated in advance on the computing cluster. Therefore, the new method for data analysis based on RSP data blocks is scalable to big data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge