Ruiying Liu
Contrast-Invariant Self-supervised Segmentation for Quantitative Placental MRI
May 30, 2025Abstract:Accurate placental segmentation is essential for quantitative analysis of the placenta. However, this task is particularly challenging in T2*-weighted placental imaging due to: (1) weak and inconsistent boundary contrast across individual echoes; (2) the absence of manual ground truth annotations for all echo times; and (3) motion artifacts across echoes caused by fetal and maternal movement. In this work, we propose a contrast-augmented segmentation framework that leverages complementary information across multi-echo T2*-weighted MRI to learn robust, contrast-invariant representations. Our method integrates: (i) masked autoencoding (MAE) for self-supervised pretraining on unlabeled multi-echo slices; (ii) masked pseudo-labeling (MPL) for unsupervised domain adaptation across echo times; and (iii) global-local collaboration to align fine-grained features with global anatomical context. We further introduce a semantic matching loss to encourage representation consistency across echoes of the same subject. Experiments on a clinical multi-echo placental MRI dataset demonstrate that our approach generalizes effectively across echo times and outperforms both single-echo and naive fusion baselines. To our knowledge, this is the first work to systematically exploit multi-echo T2*-weighted MRI for placental segmentation.
On Diffusion Process in SE-invariant Space
Mar 03, 2024
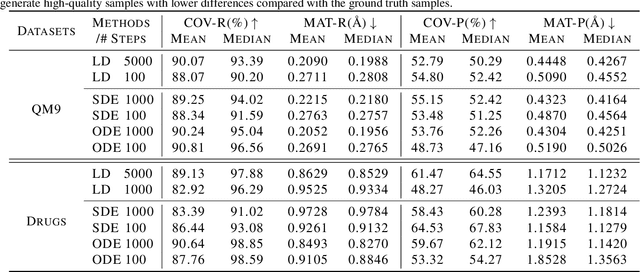
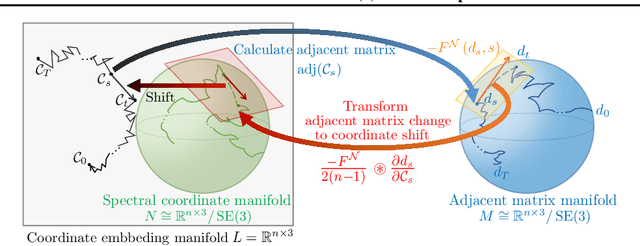
Abstract:Sampling viable 3D structures (e.g., molecules and point clouds) with SE(3)-invariance using diffusion-based models proved promising in a variety of real-world applications, wherein SE(3)-invariant properties can be naturally characterized by the inter-point distance manifold. However, due to the non-trivial geometry, we still lack a comprehensive understanding of the diffusion mechanism within such SE(3)-invariant space. This study addresses this gap by mathematically delineating the diffusion mechanism under SE(3)-invariance, via zooming into the interaction behavior between coordinates and the inter-point distance manifold through the lens of differential geometry. Upon this analysis, we propose accurate and projection-free diffusion SDE and ODE accordingly. Such formulations enable enhancing the performance and the speed of generation pathways; meanwhile offering valuable insights into other systems incorporating SE(3)-invariance.
On Accelerating Diffusion-based Molecular Conformation Generation in SE(3)-invariant Space
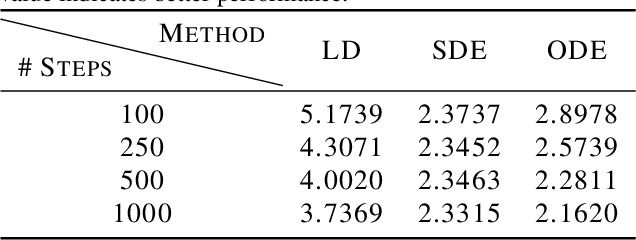
Oct 07, 2023Abstract:Diffusion-based generative models in SE(3)-invariant space have demonstrated promising performance in molecular conformation generation, but typically require solving stochastic differential equations (SDEs) with thousands of update steps. Till now, it remains unclear how to effectively accelerate this procedure explicitly in SE(3)-invariant space, which greatly hinders its wide application in the real world. In this paper, we systematically study the diffusion mechanism in SE(3)-invariant space via the lens of approximate errors induced by existing methods. Thereby, we develop more precise approximate in SE(3) in the context of projected differential equations. Theoretical analysis is further provided as well as empirical proof relating hyper-parameters with such errors. Altogether, we propose a novel acceleration scheme for generating molecular conformations in SE(3)-invariant space. Experimentally, our scheme can generate high-quality conformations with 50x--100x speedup compared to existing methods.
Molecular Conformation Generation via Shifting Scores
Sep 12, 2023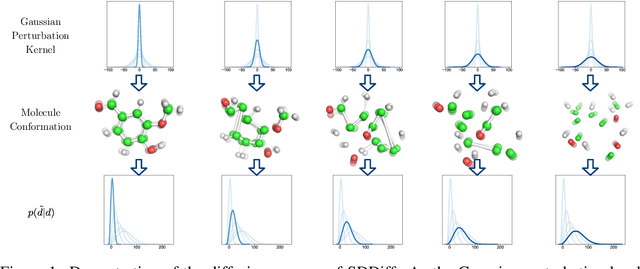
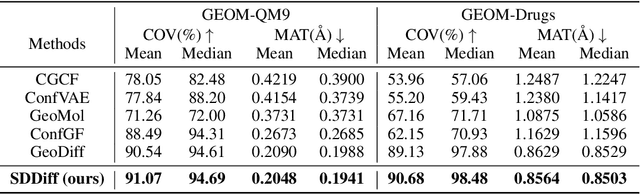
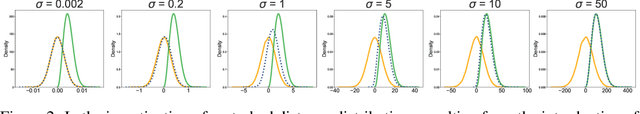
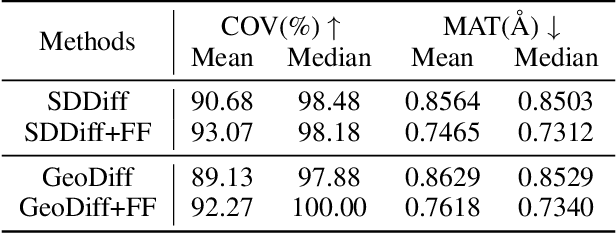
Abstract:Molecular conformation generation, a critical aspect of computational chemistry, involves producing the three-dimensional conformer geometry for a given molecule. Generating molecular conformation via diffusion requires learning to reverse a noising process. Diffusion on inter-atomic distances instead of conformation preserves SE(3)-equivalence and shows superior performance compared to alternative techniques, whereas related generative modelings are predominantly based upon heuristical assumptions. In response to this, we propose a novel molecular conformation generation approach driven by the observation that the disintegration of a molecule can be viewed as casting increasing force fields to its composing atoms, such that the distribution of the change of inter-atomic distance shifts from Gaussian to Maxwell-Boltzmann distribution. The corresponding generative modeling ensures a feasible inter-atomic distance geometry and exhibits time reversibility. Experimental results on molecular datasets demonstrate the advantages of the proposed shifting distribution compared to the state-of-the-art.
Deep Learning for Highly Accelerated Diffusion Tensor Imaging
Feb 03, 2020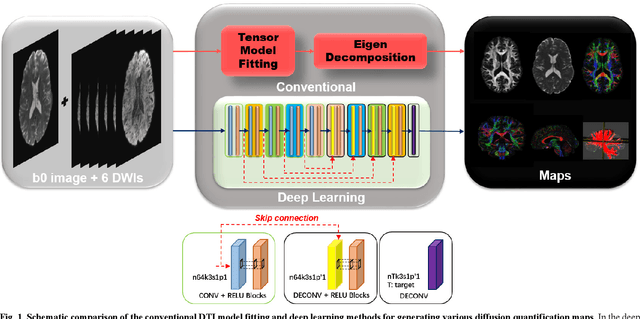
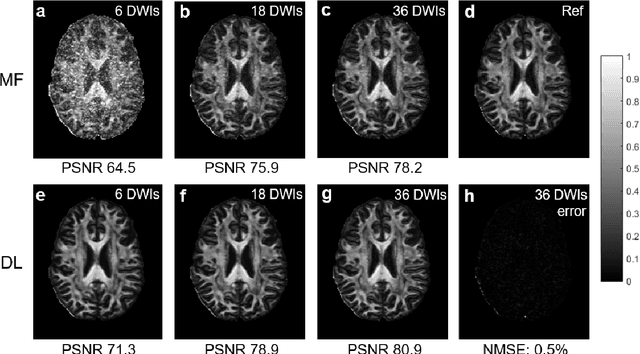
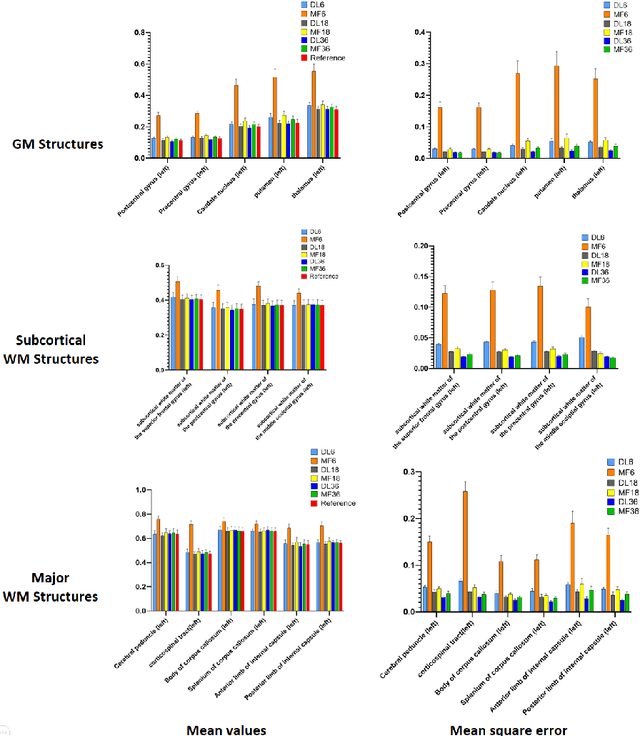

Abstract:Diffusion tensor imaging (DTI) is widely used to examine the human brain white matter structures, including their microarchitecture integrity and spatial fiber tract trajectories, with clinical applications in several neurological disorders and neurosurgical guidance. However, a major factor that prevents DTI from being incorporated in clinical routines is its long scan time due to the acquisition of a large number (typically 30 or more) of diffusion-weighted images (DWIs) required for reliable tensor estimation. Here, a deep learning-based technique is developed to obtain diffusion tensor images with only six DWIs, resulting in a significant reduction in imaging time. The method uses deep convolutional neural networks to learn the highly nonlinear relationship between DWIs and several tensor-derived maps, bypassing the conventional tensor fitting procedure, which is well known to be highly susceptible to noises in DWIs. The performance of the method was evaluated using DWI datasets from the Human Connectome Project and patients with ischemic stroke. Our results demonstrate that the proposed technique is able to generate quantitative maps of good quality fractional anisotropy (FA) and mean diffusivity (MD), as well as the fiber tractography from as few as six DWIs. The proposed method achieves a quantification error of less than 5% in all regions of interest of the brain, which is the rate of in vivo reproducibility of diffusion tensor imaging. Tractography reconstruction is also comparable to the ground truth obtained from 90 DWIs. In addition, we also demonstrate that the neural network trained on healthy volunteers can be directly applied/tested on stroke patients' DWIs data without compromising the lesion detectability. Such a significant reduction in scan time will allow inclusion of DTI into clinical routine for many potential applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge