Zach Eaton-Rosen
Imagen 3
Aug 13, 2024Abstract:We introduce Imagen 3, a latent diffusion model that generates high quality images from text prompts. We describe our quality and responsibility evaluations. Imagen 3 is preferred over other state-of-the-art (SOTA) models at the time of evaluation. In addition, we discuss issues around safety and representation, as well as methods we used to minimize the potential harm of our models.
GraphCast: Learning skillful medium-range global weather forecasting
Dec 24, 2022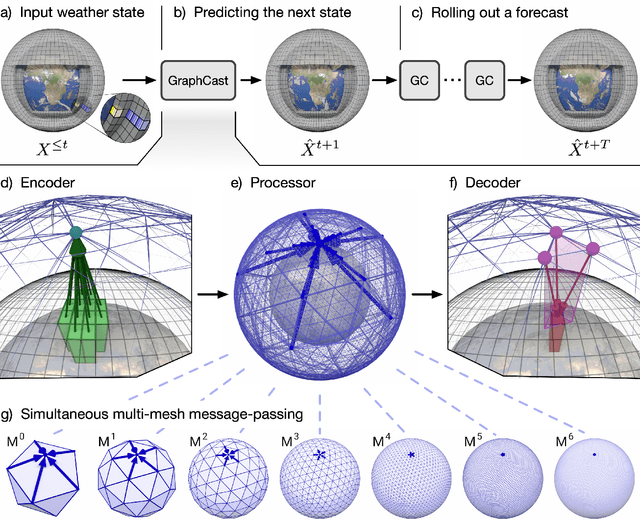
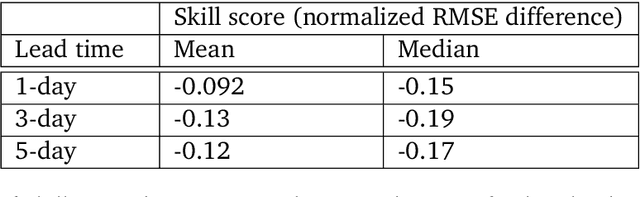
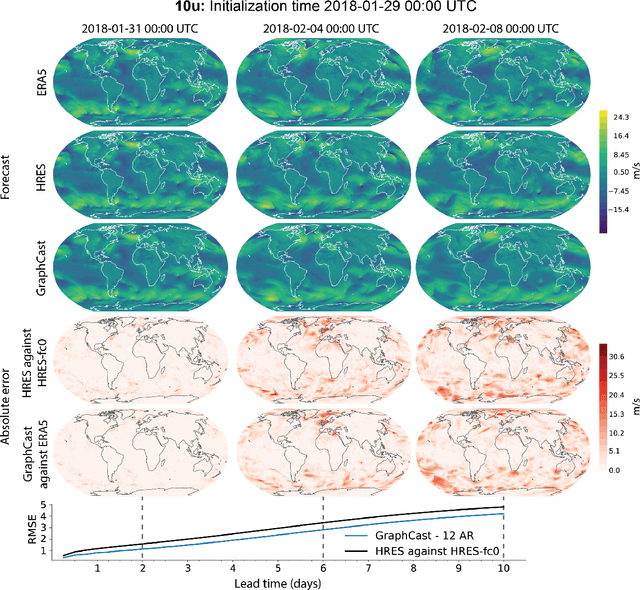
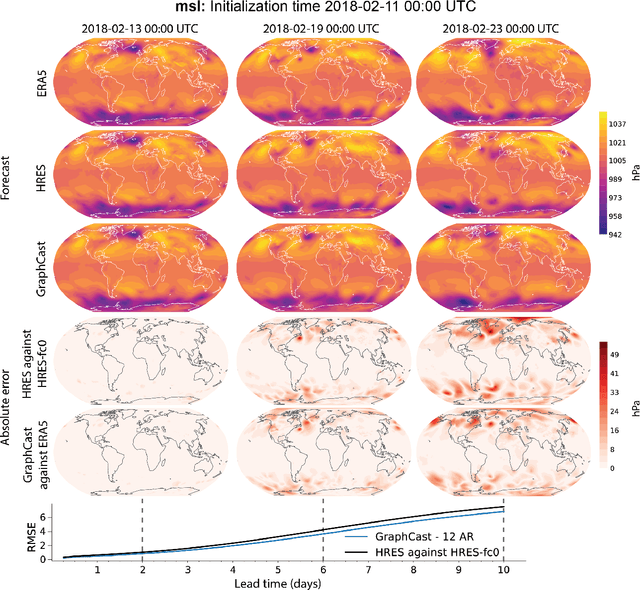
Abstract:We introduce a machine-learning (ML)-based weather simulator--called "GraphCast"--which outperforms the most accurate deterministic operational medium-range weather forecasting system in the world, as well as all previous ML baselines. GraphCast is an autoregressive model, based on graph neural networks and a novel high-resolution multi-scale mesh representation, which we trained on historical weather data from the European Centre for Medium-Range Weather Forecasts (ECMWF)'s ERA5 reanalysis archive. It can make 10-day forecasts, at 6-hour time intervals, of five surface variables and six atmospheric variables, each at 37 vertical pressure levels, on a 0.25-degree latitude-longitude grid, which corresponds to roughly 25 x 25 kilometer resolution at the equator. Our results show GraphCast is more accurate than ECMWF's deterministic operational forecasting system, HRES, on 90.0% of the 2760 variable and lead time combinations we evaluated. GraphCast also outperforms the most accurate previous ML-based weather forecasting model on 99.2% of the 252 targets it reported. GraphCast can generate a 10-day forecast (35 gigabytes of data) in under 60 seconds on Cloud TPU v4 hardware. Unlike traditional forecasting methods, ML-based forecasting scales well with data: by training on bigger, higher quality, and more recent data, the skill of the forecasts can improve. Together these results represent a key step forward in complementing and improving weather modeling with ML, open new opportunities for fast, accurate forecasting, and help realize the promise of ML-based simulation in the physical sciences.
Making EfficientNet More Efficient: Exploring Batch-Independent Normalization, Group Convolutions and Reduced Resolution Training
Jun 15, 2021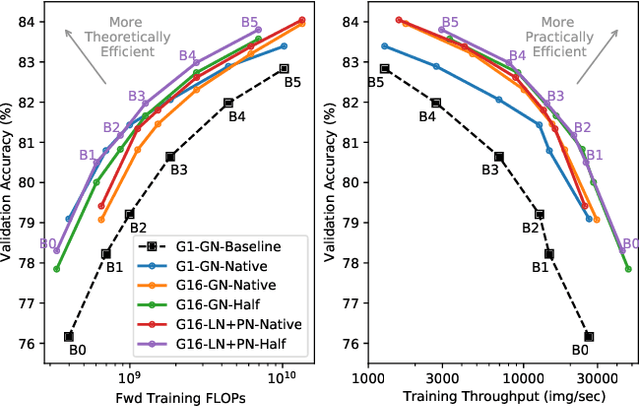

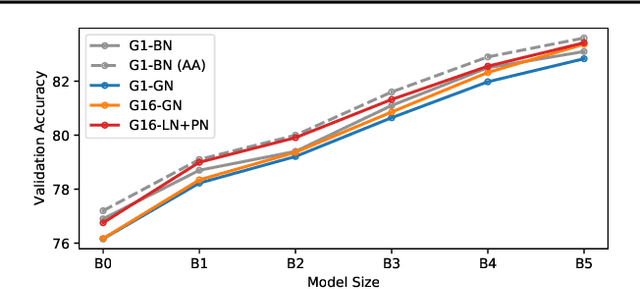
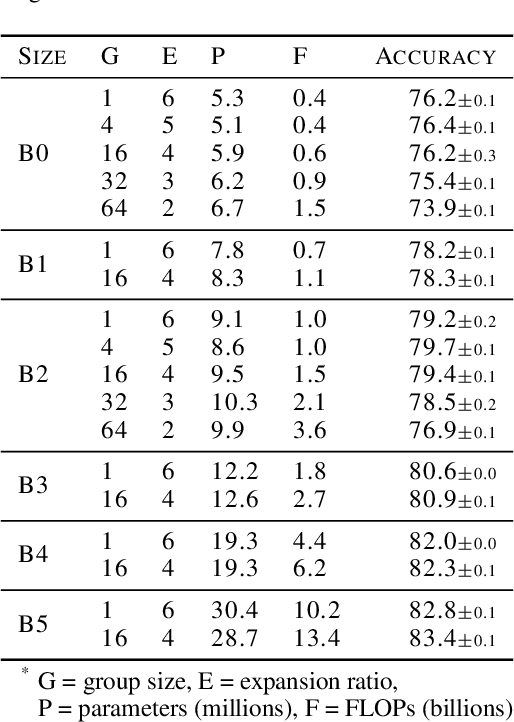
Abstract:Much recent research has been dedicated to improving the efficiency of training and inference for image classification. This effort has commonly focused on explicitly improving theoretical efficiency, often measured as ImageNet validation accuracy per FLOP. These theoretical savings have, however, proven challenging to achieve in practice, particularly on high-performance training accelerators. In this work, we focus on improving the practical efficiency of the state-of-the-art EfficientNet models on a new class of accelerator, the Graphcore IPU. We do this by extending this family of models in the following ways: (i) generalising depthwise convolutions to group convolutions; (ii) adding proxy-normalized activations to match batch normalization performance with batch-independent statistics; (iii) reducing compute by lowering the training resolution and inexpensively fine-tuning at higher resolution. We find that these three methods improve the practical efficiency for both training and inference. Our code will be made available online.
Proxy-Normalizing Activations to Match Batch Normalization while Removing Batch Dependence
Jun 11, 2021
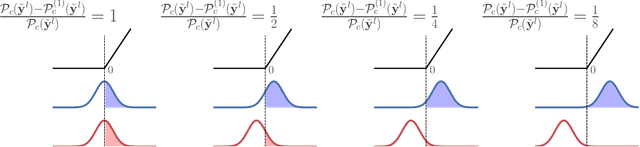

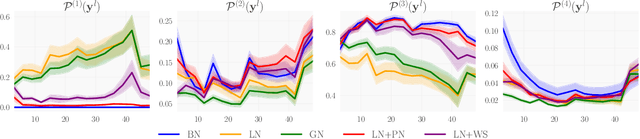
Abstract:We investigate the reasons for the performance degradation incurred with batch-independent normalization. We find that the prototypical techniques of layer normalization and instance normalization both induce the appearance of failure modes in the neural network's pre-activations: (i) layer normalization induces a collapse towards channel-wise constant functions; (ii) instance normalization induces a lack of variability in instance statistics, symptomatic of an alteration of the expressivity. To alleviate failure mode (i) without aggravating failure mode (ii), we introduce the technique "Proxy Normalization" that normalizes post-activations using a proxy distribution. When combined with layer normalization or group normalization, this batch-independent normalization emulates batch normalization's behavior and consistently matches or exceeds its performance.
Improving Neural Network Training in Low Dimensional Random Bases
Nov 09, 2020



Abstract:Stochastic Gradient Descent (SGD) has proven to be remarkably effective in optimizing deep neural networks that employ ever-larger numbers of parameters. Yet, improving the efficiency of large-scale optimization remains a vital and highly active area of research. Recent work has shown that deep neural networks can be optimized in randomly-projected subspaces of much smaller dimensionality than their native parameter space. While such training is promising for more efficient and scalable optimization schemes, its practical application is limited by inferior optimization performance. Here, we improve on recent random subspace approaches as follows: Firstly, we show that keeping the random projection fixed throughout training is detrimental to optimization. We propose re-drawing the random subspace at each step, which yields significantly better performance. We realize further improvements by applying independent projections to different parts of the network, making the approximation more efficient as network dimensionality grows. To implement these experiments, we leverage hardware-accelerated pseudo-random number generation to construct the random projections on-demand at every optimization step, allowing us to distribute the computation of independent random directions across multiple workers with shared random seeds. This yields significant reductions in memory and is up to 10 times faster for the workloads in question.
Multi-Domain Adaptation in Brain MRI through Paired Consistency and Adversarial Learning
Sep 17, 2019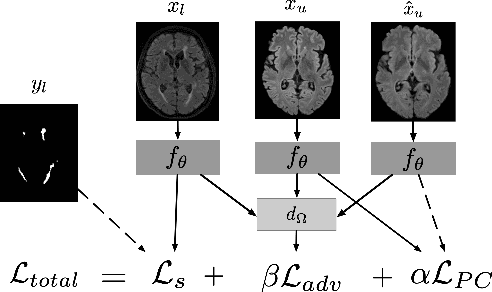
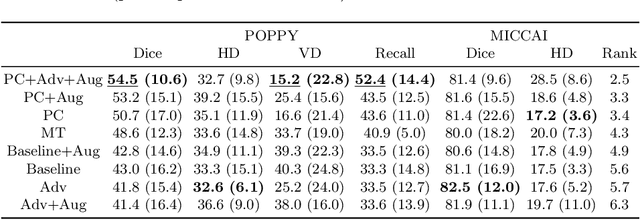
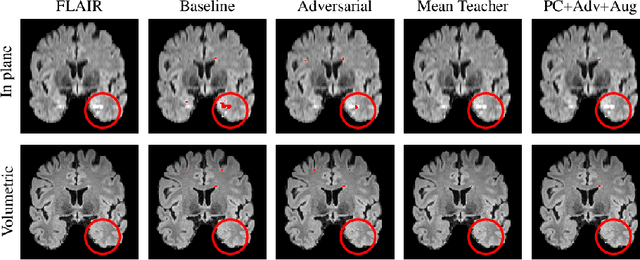
Abstract:Supervised learning algorithms trained on medical images will often fail to generalize across changes in acquisition parameters. Recent work in domain adaptation addresses this challenge and successfully leverages labeled data in a source domain to perform well on an unlabeled target domain. Inspired by recent work in semi-supervised learning we introduce a novel method to adapt from one source domain to $n$ target domains (as long as there is paired data covering all domains). Our multi-domain adaptation method utilises a consistency loss combined with adversarial learning. We provide results on white matter lesion hyperintensity segmentation from brain MRIs using the MICCAI 2017 challenge data as the source domain and two target domains. The proposed method significantly outperforms other domain adaptation baselines.
As easy as 1, 2 4? Uncertainty in counting tasks for medical imaging
Jul 25, 2019



Abstract:Counting is a fundamental task in biomedical imaging and count is an important biomarker in a number of conditions. Estimating the uncertainty in the measurement is thus vital to making definite, informed conclusions. In this paper, we first compare a range of existing methods to perform counting in medical imaging and suggest ways of deriving predictive intervals from these. We then propose and test a method for calculating intervals as an output of a multi-task network. These predictive intervals are optimised to be as narrow as possible, while also enclosing a desired percentage of the data. We demonstrate the effectiveness of this technique on histopathological cell counting and white matter hyperintensity counting. Finally, we offer insight into other areas where this technique may apply.
PIMMS: Permutation Invariant Multi-Modal Segmentation
Jul 17, 2018


Abstract:In a research context, image acquisition will often involve a pre-defined static protocol and the data will be of high quality. If we are to build applications that work in hospitals without significant operational changes in care delivery, algorithms should be designed to cope with the available data in the best possible way. In a clinical environment, imaging protocols are highly flexible, with MRI sequences commonly missing appropriate sequence labeling (e.g. T1, T2, FLAIR). To this end we introduce PIMMS, a Permutation Invariant Multi-Modal Segmentation technique that is able to perform inference over sets of MRI scans without using modality labels. We present results which show that our convolutional neural network can, in some settings, outperform a baseline model which utilizes modality labels, and achieve comparable performance otherwise.
Towards safe deep learning: accurately quantifying biomarker uncertainty in neural network predictions
Jun 22, 2018



Abstract:Automated medical image segmentation, specifically using deep learning, has shown outstanding performance in semantic segmentation tasks. However, these methods rarely quantify their uncertainty, which may lead to errors in downstream analysis. In this work we propose to use Bayesian neural networks to quantify uncertainty within the domain of semantic segmentation. We also propose a method to convert voxel-wise segmentation uncertainty into volumetric uncertainty, and calibrate the accuracy and reliability of confidence intervals of derived measurements. When applied to a tumour volume estimation application, we demonstrate that by using such modelling of uncertainty, deep learning systems can be made to report volume estimates with well-calibrated error-bars, making them safer for clinical use. We also show that the uncertainty estimates extrapolate to unseen data, and that the confidence intervals are robust in the presence of artificial noise. This could be used to provide a form of quality control and quality assurance, and may permit further adoption of deep learning tools in the clinic.
Uncertainty in multitask learning: joint representations for probabilistic MR-only radiotherapy planning
Jun 18, 2018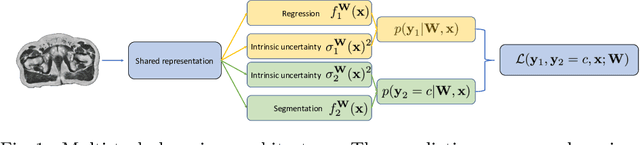
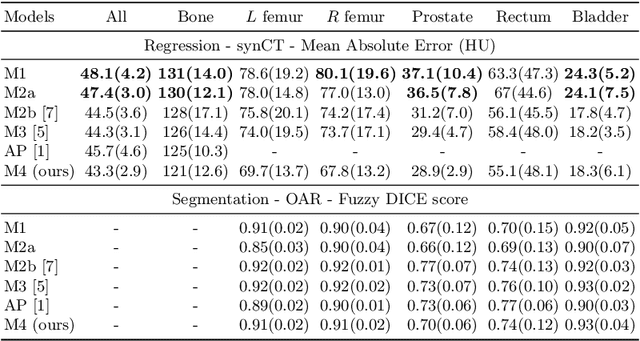
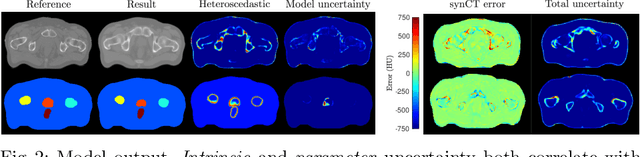
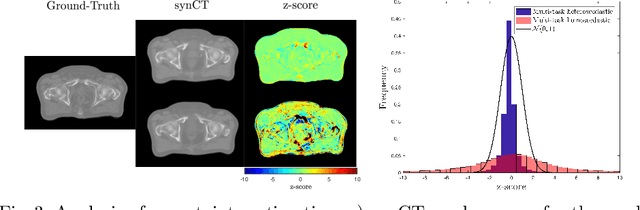
Abstract:Multi-task neural network architectures provide a mechanism that jointly integrates information from distinct sources. It is ideal in the context of MR-only radiotherapy planning as it can jointly regress a synthetic CT (synCT) scan and segment organs-at-risk (OAR) from MRI. We propose a probabilistic multi-task network that estimates: 1) intrinsic uncertainty through a heteroscedastic noise model for spatially-adaptive task loss weighting and 2) parameter uncertainty through approximate Bayesian inference. This allows sampling of multiple segmentations and synCTs that share their network representation. We test our model on prostate cancer scans and show that it produces more accurate and consistent synCTs with a better estimation in the variance of the errors, state of the art results in OAR segmentation and a methodology for quality assurance in radiotherapy treatment planning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge