Mads Nielsen
Deep Learning-Based Regional White Matter Hyperintensity Mapping as a Robust Biomarker for Alzheimer's Disease
Nov 18, 2025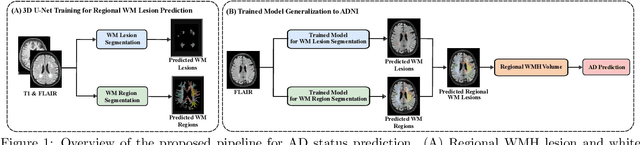
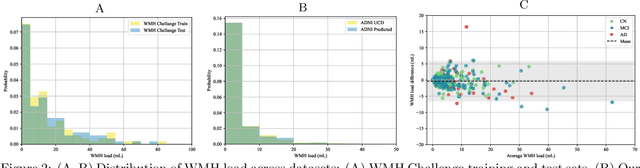
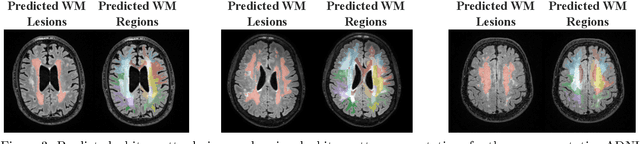
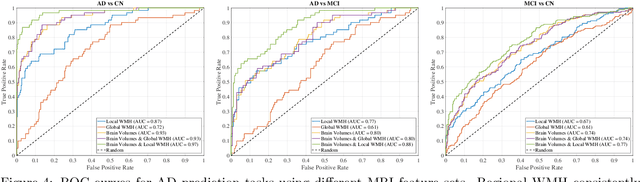
Abstract:White matter hyperintensities (WMH) are key imaging markers in cognitive aging, Alzheimer's disease (AD), and related dementias. Although automated methods for WMH segmentation have advanced, most provide only global lesion load and overlook their spatial distribution across distinct white matter regions. We propose a deep learning framework for robust WMH segmentation and localization, evaluated across public datasets and an independent Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort. Our results show that the predicted lesion loads are in line with the reference WMH estimates, confirming the robustness to variations in lesion load, acquisition, and demographics. Beyond accurate segmentation, we quantify WMH load within anatomically defined regions and combine these measures with brain structure volumes to assess diagnostic value. Regional WMH volumes consistently outperform global lesion burden for disease classification, and integration with brain atrophy metrics further improves performance, reaching area under the curve (AUC) values up to 0.97. Several spatially distinct regions, particularly within anterior white matter tracts, are reproducibly associated with diagnostic status, indicating localized vulnerability in AD. These results highlight the added value of regional WMH quantification. Incorporating localized lesion metrics alongside atrophy markers may enhance early diagnosis and stratification in neurodegenerative disorders.
General Methods Make Great Domain-specific Foundation Models: A Case-study on Fetal Ultrasound
Jun 24, 2025Abstract:With access to large-scale, unlabeled medical datasets, researchers are confronted with two questions: Should they attempt to pretrain a custom foundation model on this medical data, or use transfer-learning from an existing generalist model? And, if a custom model is pretrained, are novel methods required? In this paper we explore these questions by conducting a case-study, in which we train a foundation model on a large regional fetal ultrasound dataset of 2M images. By selecting the well-established DINOv2 method for pretraining, we achieve state-of-the-art results on three fetal ultrasound datasets, covering data from different countries, classification, segmentation, and few-shot tasks. We compare against a series of models pretrained on natural images, ultrasound images, and supervised baselines. Our results demonstrate two key insights: (i) Pretraining on custom data is worth it, even if smaller models are trained on less data, as scaling in natural image pretraining does not translate to ultrasound performance. (ii) Well-tuned methods from computer vision are making it feasible to train custom foundation models for a given medical domain, requiring no hyperparameter tuning and little methodological adaptation. Given these findings, we argue that a bias towards methodological innovation should be avoided when developing domain specific foundation models under common computational resource constraints.
A large-scale heterogeneous 3D magnetic resonance brain imaging dataset for self-supervised learning
Jun 17, 2025Abstract:We present FOMO60K, a large-scale, heterogeneous dataset of 60,529 brain Magnetic Resonance Imaging (MRI) scans from 13,900 sessions and 11,187 subjects, aggregated from 16 publicly available sources. The dataset includes both clinical- and research-grade images, multiple MRI sequences, and a wide range of anatomical and pathological variability, including scans with large brain anomalies. Minimal preprocessing was applied to preserve the original image characteristics while reducing barriers to entry for new users. Accompanying code for self-supervised pretraining and finetuning is provided. FOMO60K is intended to support the development and benchmarking of self-supervised learning methods in medical imaging at scale.
A General Purpose Spectral Foundational Model for Both Proximal and Remote Sensing Spectral Imaging
Mar 03, 2025



Abstract:Spectral imaging data acquired via multispectral and hyperspectral cameras can have hundreds of channels, where each channel records the reflectance at a specific wavelength and bandwidth. Time and resource constraints limit our ability to collect large spectral datasets, making it difficult to build and train predictive models from scratch. In the RGB domain, we can often alleviate some of the limitations of smaller datasets by using pretrained foundational models as a starting point. However, most existing foundation models are pretrained on large datasets of 3-channel RGB images, severely limiting their effectiveness when used with spectral imaging data. The few spectral foundation models that do exist usually have one of two limitations: (1) they are built and trained only on remote sensing data limiting their application in proximal spectral imaging, (2) they utilize the more widely available multispectral imaging datasets with less than 15 channels restricting their use with hundred-channel hyperspectral images. To alleviate these issues, we propose a large-scale foundational model and dataset built upon the masked autoencoder architecture that takes advantage of spectral channel encoding, spatial-spectral masking and ImageNet pretraining for an adaptable and robust model for downstream spectral imaging tasks.
Revisiting CLIP: Efficient Alignment of 3D MRI and Tabular Data using Domain-Specific Foundation Models
Jan 23, 2025



Abstract:Multi-modal models require aligned, shared embedding spaces. However, common CLIP-based approaches need large amounts of samples and do not natively support 3D or tabular data, both of which are crucial in the medical domain. To address these issues, we revisit CLIP-style alignment by training a domain-specific 3D foundation model as an image encoder and demonstrate that modality alignment is feasible with only 62 MRI scans. Our approach is enabled by a simple embedding accumulation strategy required for training in 3D, which scales the amount of negative pairs across batches in order to stabilize training. We perform a thorough evaluation of various design choices, including the choice of backbone and loss functions, and evaluate the proposed methodology on zero-shot classification and image-retrieval tasks. While zero-shot image-retrieval remains challenging, zero-shot classification results demonstrate that the proposed approach can meaningfully align the representations of 3D MRI with tabular data.
Assessing the Efficacy of Classical and Deep Neuroimaging Biomarkers in Early Alzheimer's Disease Diagnosis
Oct 31, 2024



Abstract:Alzheimer's disease (AD) is the leading cause of dementia, and its early detection is crucial for effective intervention, yet current diagnostic methods often fall short in sensitivity and specificity. This study aims to detect significant indicators of early AD by extracting and integrating various imaging biomarkers, including radiomics, hippocampal texture descriptors, cortical thickness measurements, and deep learning features. We analyze structural magnetic resonance imaging (MRI) scans from the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohorts, utilizing comprehensive image analysis and machine learning techniques. Our results show that combining multiple biomarkers significantly improves detection accuracy. Radiomics and texture features emerged as the most effective predictors for early AD, achieving AUCs of 0.88 and 0.72 for AD and MCI detection, respectively. Although deep learning features proved to be less effective than traditional approaches, incorporating age with other biomarkers notably enhanced MCI detection performance. Additionally, our findings emphasize the continued importance of classical imaging biomarkers in the face of modern deep-learning approaches, providing a robust framework for early AD diagnosis.
Unsupervised Detection of Fetal Brain Anomalies using Denoising Diffusion Models
Aug 07, 2024Abstract:Congenital malformations of the brain are among the most common fetal abnormalities that impact fetal development. Previous anomaly detection methods on ultrasound images are based on supervised learning, rely on manual annotations, and risk missing underrepresented categories. In this work, we frame fetal brain anomaly detection as an unsupervised task using diffusion models. To this end, we employ an inpainting-based Noise Agnostic Anomaly Detection approach that identifies the abnormality using diffusion-reconstructed fetal brain images from multiple noise levels. Our approach only requires normal fetal brain ultrasound images for training, addressing the limited availability of abnormal data. Our experiments on a real-world clinical dataset show the potential of using unsupervised methods for fetal brain anomaly detection. Additionally, we comprehensively evaluate how different noise types affect diffusion models in the fetal anomaly detection domain.
AMAES: Augmented Masked Autoencoder Pretraining on Public Brain MRI Data for 3D-Native Segmentation
Aug 01, 2024Abstract:This study investigates the impact of self-supervised pretraining of 3D semantic segmentation models on a large-scale, domain-specific dataset. We introduce BRAINS-45K, a dataset of 44,756 brain MRI volumes from public sources, the largest public dataset available, and revisit a number of design choices for pretraining modern segmentation architectures by simplifying and optimizing state-of-the-art methods, and combining them with a novel augmentation strategy. The resulting AMAES framework is based on masked-image-modeling and intensity-based augmentation reversal and balances memory usage, runtime, and finetuning performance. Using the popular U-Net and the recent MedNeXt architecture as backbones, we evaluate the effect of pretraining on three challenging downstream tasks, covering single-sequence, low-resource settings, and out-of-domain generalization. The results highlight that pretraining on the proposed dataset with AMAES significantly improves segmentation performance in the majority of evaluated cases, and that it is beneficial to pretrain the model with augmentations, despite pretraing on a large-scale dataset. Code and model checkpoints for reproducing results, as well as the BRAINS-45K dataset are available at \url{https://github.com/asbjrnmunk/amaes}.
Yucca: A Deep Learning Framework For Medical Image Analysis
Jul 29, 2024


Abstract:Medical image analysis using deep learning frameworks has advanced healthcare by automating complex tasks, but many existing frameworks lack flexibility, modularity, and user-friendliness. To address these challenges, we introduce Yucca, an open-source AI framework available at https://github.com/Sllambias/yucca, designed specifically for medical imaging applications and built on PyTorch and PyTorch Lightning. Yucca features a three-tiered architecture: Functional, Modules, and Pipeline, providing a comprehensive and customizable solution. Evaluated across diverse tasks such as cerebral microbleeds detection, white matter hyperintensity segmentation, and hippocampus segmentation, Yucca achieves state-of-the-art results, demonstrating its robustness and versatility. Yucca offers a powerful, flexible, and user-friendly platform for medical image analysis, inviting community contributions to advance its capabilities and impact.
CORE-BEHRT: A Carefully Optimized and Rigorously Evaluated BEHRT
Apr 24, 2024Abstract:BERT-based models for Electronic Health Records (EHR) have surged in popularity following the release of BEHRT and Med-BERT. Subsequent models have largely built on these foundations despite the fundamental design choices of these pioneering models remaining underexplored. To address this issue, we introduce CORE-BEHRT, a Carefully Optimized and Rigorously Evaluated BEHRT. Through incremental optimization, we isolate the sources of improvement for key design choices, giving us insights into the effect of data representation and individual technical components on performance. Evaluating this across a set of generic tasks (death, pain treatment, and general infection), we showed that improving data representation can increase the average downstream performance from 0.785 to 0.797 AUROC, primarily when including medication and timestamps. Improving the architecture and training protocol on top of this increased average downstream performance to 0.801 AUROC. We then demonstrated the consistency of our optimization through a rigorous evaluation across 25 diverse clinical prediction tasks. We observed significant performance increases in 17 out of 25 tasks and improvements in 24 tasks, highlighting the generalizability of our findings. Our findings provide a strong foundation for future work and aim to increase the trustworthiness of BERT-based EHR models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge