Manxi Lin
U2-BENCH: Benchmarking Large Vision-Language Models on Ultrasound Understanding
May 23, 2025Abstract:Ultrasound is a widely-used imaging modality critical to global healthcare, yet its interpretation remains challenging due to its varying image quality on operators, noises, and anatomical structures. Although large vision-language models (LVLMs) have demonstrated impressive multimodal capabilities across natural and medical domains, their performance on ultrasound remains largely unexplored. We introduce U2-BENCH, the first comprehensive benchmark to evaluate LVLMs on ultrasound understanding across classification, detection, regression, and text generation tasks. U2-BENCH aggregates 7,241 cases spanning 15 anatomical regions and defines 8 clinically inspired tasks, such as diagnosis, view recognition, lesion localization, clinical value estimation, and report generation, across 50 ultrasound application scenarios. We evaluate 20 state-of-the-art LVLMs, both open- and closed-source, general-purpose and medical-specific. Our results reveal strong performance on image-level classification, but persistent challenges in spatial reasoning and clinical language generation. U2-BENCH establishes a rigorous and unified testbed to assess and accelerate LVLM research in the uniquely multimodal domain of medical ultrasound imaging.
Unsupervised Detection of Fetal Brain Anomalies using Denoising Diffusion Models
Aug 07, 2024Abstract:Congenital malformations of the brain are among the most common fetal abnormalities that impact fetal development. Previous anomaly detection methods on ultrasound images are based on supervised learning, rely on manual annotations, and risk missing underrepresented categories. In this work, we frame fetal brain anomaly detection as an unsupervised task using diffusion models. To this end, we employ an inpainting-based Noise Agnostic Anomaly Detection approach that identifies the abnormality using diffusion-reconstructed fetal brain images from multiple noise levels. Our approach only requires normal fetal brain ultrasound images for training, addressing the limited availability of abnormal data. Our experiments on a real-world clinical dataset show the potential of using unsupervised methods for fetal brain anomaly detection. Additionally, we comprehensively evaluate how different noise types affect diffusion models in the fetal anomaly detection domain.
Incorporating Clinical Guidelines through Adapting Multi-modal Large Language Model for Prostate Cancer PI-RADS Scoring
May 14, 2024Abstract:The Prostate Imaging Reporting and Data System (PI-RADS) is pivotal in the diagnosis of clinically significant prostate cancer through MRI imaging. Current deep learning-based PI-RADS scoring methods often lack the incorporation of essential PI-RADS clinical guidelines~(PICG) utilized by radiologists, potentially compromising scoring accuracy. This paper introduces a novel approach that adapts a multi-modal large language model (MLLM) to incorporate PICG into PI-RADS scoring without additional annotations and network parameters. We present a two-stage fine-tuning process aimed at adapting MLLMs originally trained on natural images to the MRI data domain while effectively integrating the PICG. In the first stage, we develop a domain adapter layer specifically tailored for processing 3D MRI image inputs and design the MLLM instructions to differentiate MRI modalities effectively. In the second stage, we translate PICG into guiding instructions for the model to generate PICG-guided image features. Through feature distillation, we align scoring network features with the PICG-guided image feature, enabling the scoring network to effectively incorporate the PICG information. We develop our model on a public dataset and evaluate it in a real-world challenging in-house dataset. Experimental results demonstrate that our approach improves the performance of current scoring networks.
Tri-modal Confluence with Temporal Dynamics for Scene Graph Generation in Operating Rooms
Apr 14, 2024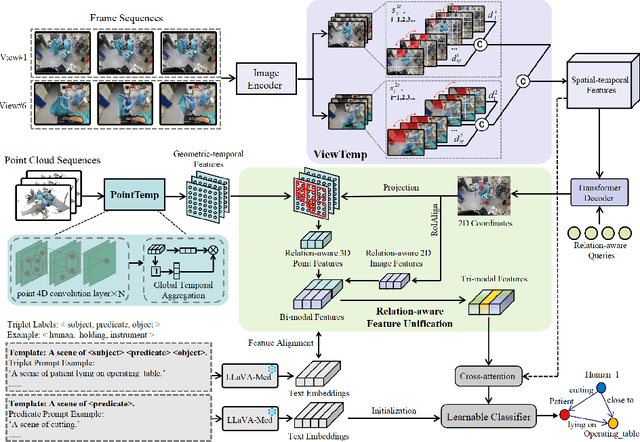
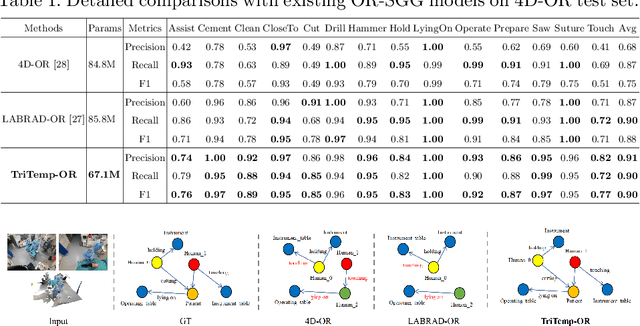
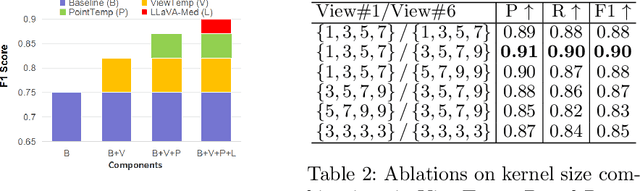

Abstract:A comprehensive understanding of surgical scenes allows for monitoring of the surgical process, reducing the occurrence of accidents and enhancing efficiency for medical professionals. Semantic modeling within operating rooms, as a scene graph generation (SGG) task, is challenging since it involves consecutive recognition of subtle surgical actions over prolonged periods. To address this challenge, we propose a Tri-modal (i.e., images, point clouds, and language) confluence with Temporal dynamics framework, termed TriTemp-OR. Diverging from previous approaches that integrated temporal information via memory graphs, our method embraces two advantages: 1) we directly exploit bi-modal temporal information from the video streaming for hierarchical feature interaction, and 2) the prior knowledge from Large Language Models (LLMs) is embedded to alleviate the class-imbalance problem in the operating theatre. Specifically, our model performs temporal interactions across 2D frames and 3D point clouds, including a scale-adaptive multi-view temporal interaction (ViewTemp) and a geometric-temporal point aggregation (PointTemp). Furthermore, we transfer knowledge from the biomedical LLM, LLaVA-Med, to deepen the comprehension of intraoperative relations. The proposed TriTemp-OR enables the aggregation of tri-modal features through relation-aware unification to predict relations so as to generate scene graphs. Experimental results on the 4D-OR benchmark demonstrate the superior performance of our model for long-term OR streaming.
Deployment of Deep Learning Model in Real World Clinical Setting: A Case Study in Obstetric Ultrasound
Mar 22, 2024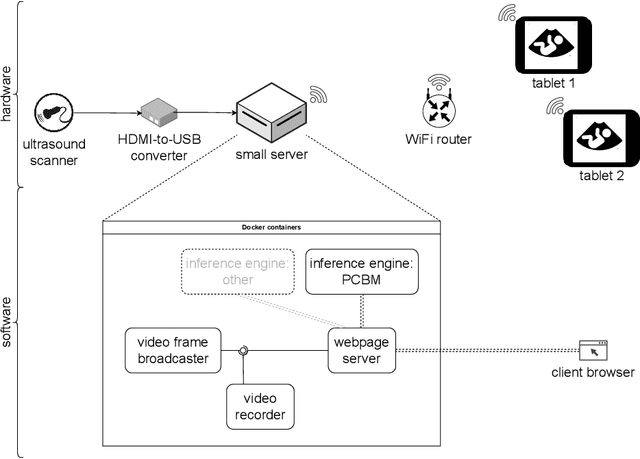

Abstract:Despite the rapid development of AI models in medical image analysis, their validation in real-world clinical settings remains limited. To address this, we introduce a generic framework designed for deploying image-based AI models in such settings. Using this framework, we deployed a trained model for fetal ultrasound standard plane detection, and evaluated it in real-time sessions with both novice and expert users. Feedback from these sessions revealed that while the model offers potential benefits to medical practitioners, the need for navigational guidance was identified as a key area for improvement. These findings underscore the importance of early deployment of AI models in real-world settings, leading to insights that can guide the refinement of the model and system based on actual user feedback.
Diffusion-based Iterative Counterfactual Explanations for Fetal Ultrasound Image Quality Assessment
Mar 13, 2024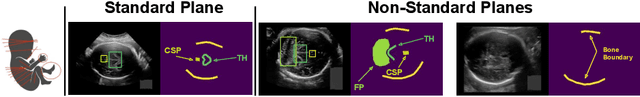



Abstract:Obstetric ultrasound image quality is crucial for accurate diagnosis and monitoring of fetal health. However, producing high-quality standard planes is difficult, influenced by the sonographer's expertise and factors like the maternal BMI or the fetus dynamics. In this work, we propose using diffusion-based counterfactual explainable AI to generate realistic high-quality standard planes from low-quality non-standard ones. Through quantitative and qualitative evaluation, we demonstrate the effectiveness of our method in producing plausible counterfactuals of increased quality. This shows future promise both for enhancing training of clinicians by providing visual feedback, as well as for improving image quality and, consequently, downstream diagnosis and monitoring.
Shortcut Learning in Medical Image Segmentation
Mar 11, 2024Abstract:Shortcut learning is a phenomenon where machine learning models prioritize learning simple, potentially misleading cues from data that do not generalize well beyond the training set. While existing research primarily investigates this in the realm of image classification, this study extends the exploration of shortcut learning into medical image segmentation. We demonstrate that clinical annotations such as calipers, and the combination of zero-padded convolutions and center-cropped training sets in the dataset can inadvertently serve as shortcuts, impacting segmentation accuracy. We identify and evaluate the shortcut learning on two different but common medical image segmentation tasks. In addition, we suggest strategies to mitigate the influence of shortcut learning and improve the generalizability of the segmentation models. By uncovering the presence and implications of shortcuts in medical image segmentation, we provide insights and methodologies for evaluating and overcoming this pervasive challenge and call for attention in the community for shortcuts in segmentation.
S^2Former-OR: Single-Stage Bimodal Transformer for Scene Graph Generation in OR
Feb 22, 2024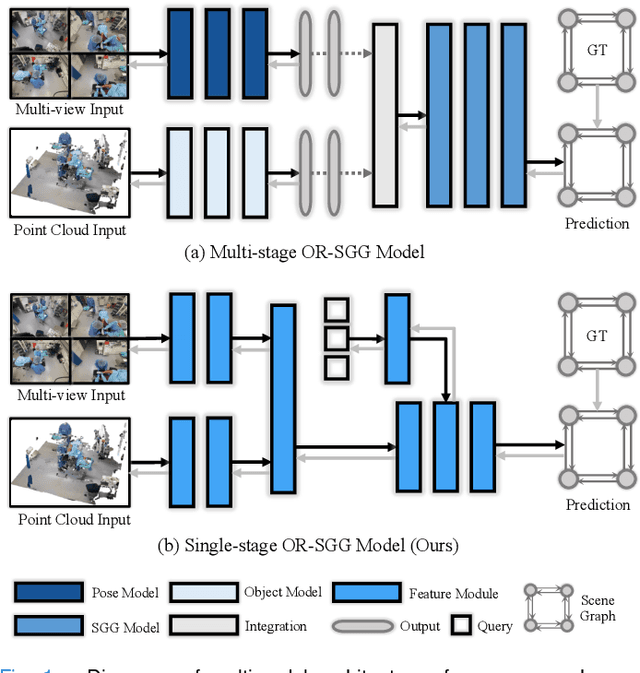
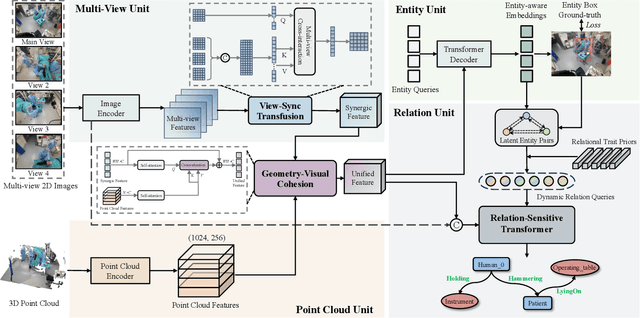
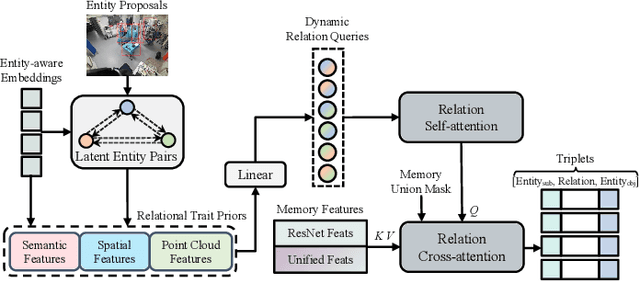
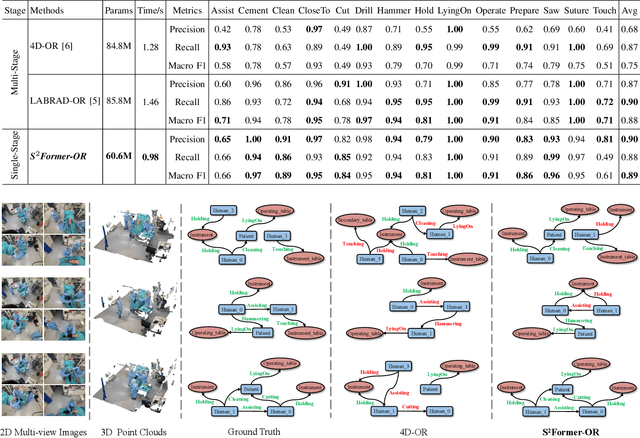
Abstract:Scene graph generation (SGG) of surgical procedures is crucial in enhancing holistically cognitive intelligence in the operating room (OR). However, previous works have primarily relied on the multi-stage learning that generates semantic scene graphs dependent on intermediate processes with pose estimation and object detection, which may compromise model efficiency and efficacy, also impose extra annotation burden. In this study, we introduce a novel single-stage bimodal transformer framework for SGG in the OR, termed S^2Former-OR, aimed to complementally leverage multi-view 2D scenes and 3D point clouds for SGG in an end-to-end manner. Concretely, our model embraces a View-Sync Transfusion scheme to encourage multi-view visual information interaction. Concurrently, a Geometry-Visual Cohesion operation is designed to integrate the synergic 2D semantic features into 3D point cloud features. Moreover, based on the augmented feature, we propose a novel relation-sensitive transformer decoder that embeds dynamic entity-pair queries and relational trait priors, which enables the direct prediction of entity-pair relations for graph generation without intermediate steps. Extensive experiments have validated the superior SGG performance and lower computational cost of S^2Former-OR on 4D-OR benchmark, compared with current OR-SGG methods, e.g., 3% Precision increase and 24.2M reduction in model parameters. We further compared our method with generic single-stage SGG methods with broader metrics for a comprehensive evaluation, with consistently better performance achieved. The code will be made available.
Learning semantic image quality for fetal ultrasound from noisy ranking annotation
Feb 13, 2024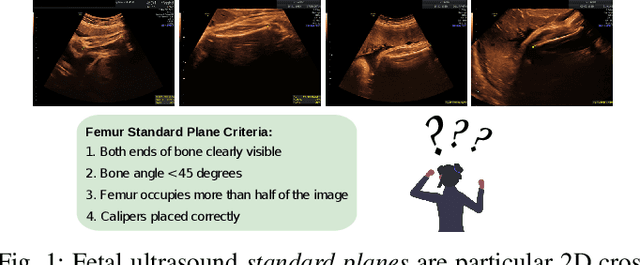
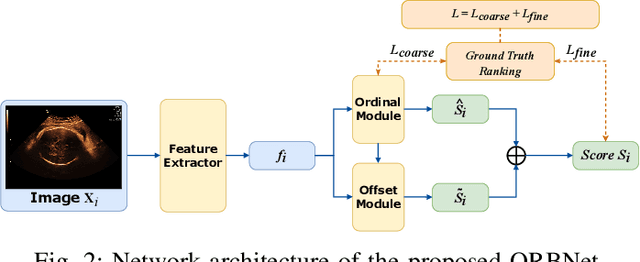
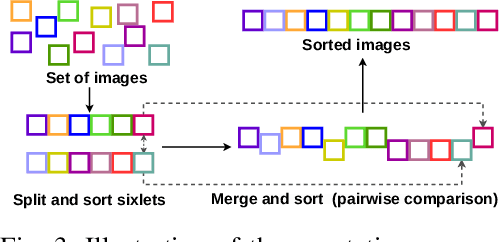
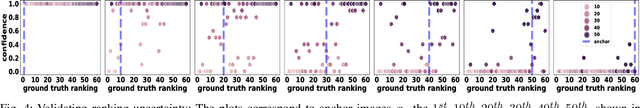
Abstract:We introduce the notion of semantic image quality for applications where image quality relies on semantic requirements. Working in fetal ultrasound, where ranking is challenging and annotations are noisy, we design a robust coarse-to-fine model that ranks images based on their semantic image quality and endow our predicted rankings with an uncertainty estimate. To annotate rankings on training data, we design an efficient ranking annotation scheme based on the merge sort algorithm. Finally, we compare our ranking algorithm to a number of state-of-the-art ranking algorithms on a challenging fetal ultrasound quality assessment task, showing the superior performance of our method on the majority of rank correlation metrics.
An Automatic Guidance and Quality Assessment System for Doppler Imaging of Umbilical Artery
Apr 11, 2023Abstract:In fetal ultrasound screening, Doppler images on the umbilical artery (UA) are important for monitoring blood supply through the umbilical cord. However, to capture UA Doppler images, a number of steps need to be done correctly: placing the gate at a proper location in the ultrasound image to obtain blood flow waveforms, and judging the Doppler waveform quality. Both of these rely on the operator's experience. The shortage of experienced sonographers thus creates a demand for machine assistance. We propose an automatic system to fill this gap. Using a modified Faster R-CNN we obtain an algorithm that suggests Doppler flow gate locations. We subsequently assess the Doppler waveform quality. We validate the proposed system on 657 scans from a national ultrasound screening database. The experimental results demonstrate that our system is useful in guiding operators for UA Doppler image capture and quality assessment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge