Kamil Mikolaj
General Methods Make Great Domain-specific Foundation Models: A Case-study on Fetal Ultrasound
Jun 24, 2025Abstract:With access to large-scale, unlabeled medical datasets, researchers are confronted with two questions: Should they attempt to pretrain a custom foundation model on this medical data, or use transfer-learning from an existing generalist model? And, if a custom model is pretrained, are novel methods required? In this paper we explore these questions by conducting a case-study, in which we train a foundation model on a large regional fetal ultrasound dataset of 2M images. By selecting the well-established DINOv2 method for pretraining, we achieve state-of-the-art results on three fetal ultrasound datasets, covering data from different countries, classification, segmentation, and few-shot tasks. We compare against a series of models pretrained on natural images, ultrasound images, and supervised baselines. Our results demonstrate two key insights: (i) Pretraining on custom data is worth it, even if smaller models are trained on less data, as scaling in natural image pretraining does not translate to ultrasound performance. (ii) Well-tuned methods from computer vision are making it feasible to train custom foundation models for a given medical domain, requiring no hyperparameter tuning and little methodological adaptation. Given these findings, we argue that a bias towards methodological innovation should be avoided when developing domain specific foundation models under common computational resource constraints.
Shortcut Learning in Medical Image Segmentation
Mar 11, 2024Abstract:Shortcut learning is a phenomenon where machine learning models prioritize learning simple, potentially misleading cues from data that do not generalize well beyond the training set. While existing research primarily investigates this in the realm of image classification, this study extends the exploration of shortcut learning into medical image segmentation. We demonstrate that clinical annotations such as calipers, and the combination of zero-padded convolutions and center-cropped training sets in the dataset can inadvertently serve as shortcuts, impacting segmentation accuracy. We identify and evaluate the shortcut learning on two different but common medical image segmentation tasks. In addition, we suggest strategies to mitigate the influence of shortcut learning and improve the generalizability of the segmentation models. By uncovering the presence and implications of shortcuts in medical image segmentation, we provide insights and methodologies for evaluating and overcoming this pervasive challenge and call for attention in the community for shortcuts in segmentation.
Removing confounding information from fetal ultrasound images
Mar 24, 2023



Abstract:Confounding information in the form of text or markings embedded in medical images can severely affect the training of diagnostic deep learning algorithms. However, data collected for clinical purposes often have such markings embedded in them. In dermatology, known examples include drawings or rulers that are overrepresented in images of malignant lesions. In this paper, we encounter text and calipers placed on the images found in national databases containing fetal screening ultrasound scans, which correlate with standard planes to be predicted. In order to utilize the vast amounts of data available in these databases, we develop and validate a series of methods for minimizing the confounding effects of embedded text and calipers on deep learning algorithms designed for ultrasound, using standard plane classification as a test case.
Generalisability of deep learning models in low-resource imaging settings: A fetal ultrasound study in 5 African countries
Sep 20, 2022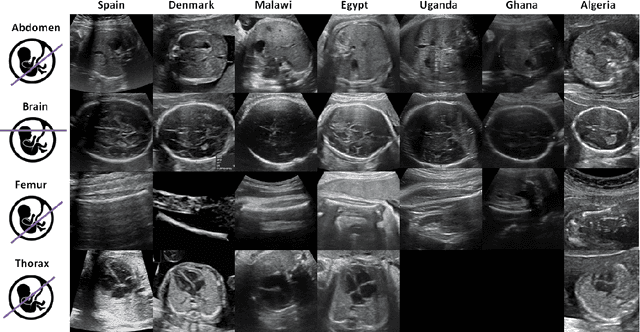
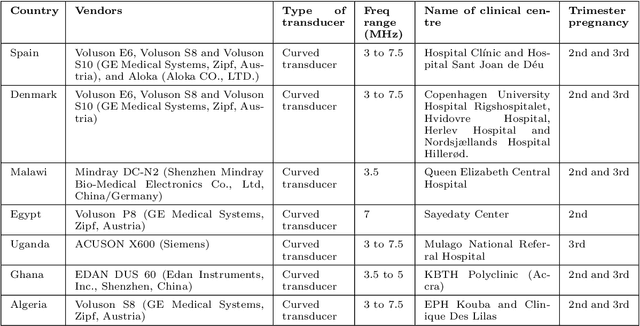
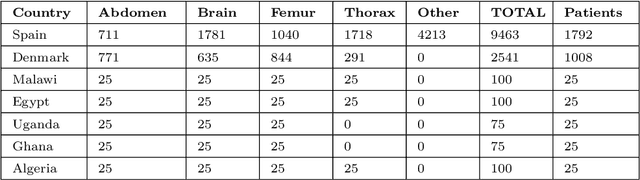
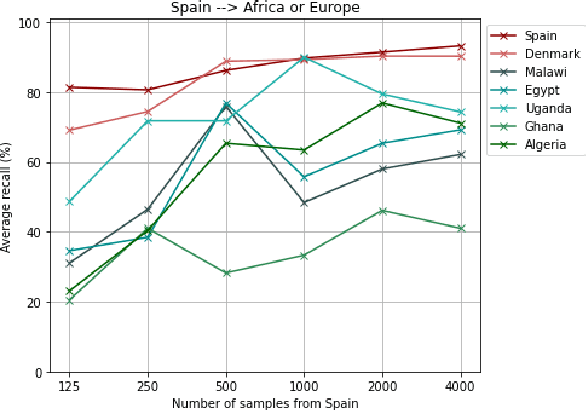
Abstract:Most artificial intelligence (AI) research have concentrated in high-income countries, where imaging data, IT infrastructures and clinical expertise are plentiful. However, slower progress has been made in limited-resource environments where medical imaging is needed. For example, in Sub-Saharan Africa the rate of perinatal mortality is very high due to limited access to antenatal screening. In these countries, AI models could be implemented to help clinicians acquire fetal ultrasound planes for diagnosis of fetal abnormalities. So far, deep learning models have been proposed to identify standard fetal planes, but there is no evidence of their ability to generalise in centres with limited access to high-end ultrasound equipment and data. This work investigates different strategies to reduce the domain-shift effect for a fetal plane classification model trained on a high-resource clinical centre and transferred to a new low-resource centre. To that end, a classifier trained with 1,792 patients from Spain is first evaluated on a new centre in Denmark in optimal conditions with 1,008 patients and is later optimised to reach the same performance in five African centres (Egypt, Algeria, Uganda, Ghana and Malawi) with 25 patients each. The results show that a transfer learning approach can be a solution to integrate small-size African samples with existing large-scale databases in developed countries. In particular, the model can be re-aligned and optimised to boost the performance on African populations by increasing the recall to $0.92 \pm 0.04$ and at the same time maintaining a high precision across centres. This framework shows promise for building new AI models generalisable across clinical centres with limited data acquired in challenging and heterogeneous conditions and calls for further research to develop new solutions for usability of AI in countries with less resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge