Jordina Torrents-Barrena
Invariance Measures for Neural Networks
Oct 26, 2023Abstract:Invariances in neural networks are useful and necessary for many tasks. However, the representation of the invariance of most neural network models has not been characterized. We propose measures to quantify the invariance of neural networks in terms of their internal representation. The measures are efficient and interpretable, and can be applied to any neural network model. They are also more sensitive to invariance than previously defined measures. We validate the measures and their properties in the domain of affine transformations and the CIFAR10 and MNIST datasets, including their stability and interpretability. Using the measures, we perform a first analysis of CNN models and show that their internal invariance is remarkably stable to random weight initializations, but not to changes in dataset or transformation. We believe the measures will enable new avenues of research in invariance representation.
Generalisability of deep learning models in low-resource imaging settings: A fetal ultrasound study in 5 African countries
Sep 20, 2022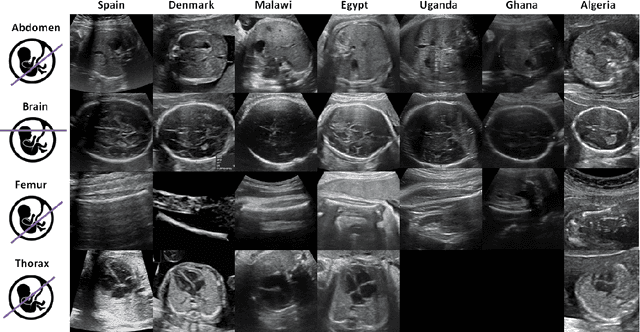
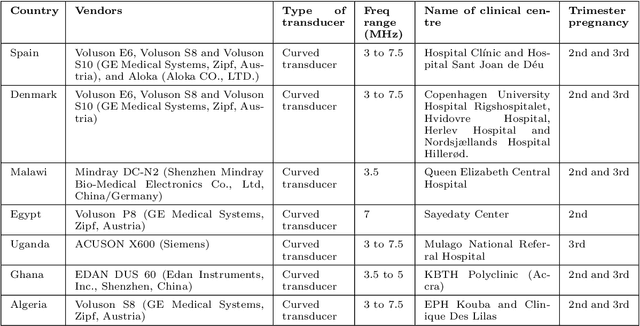
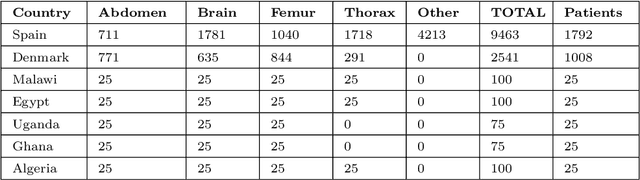
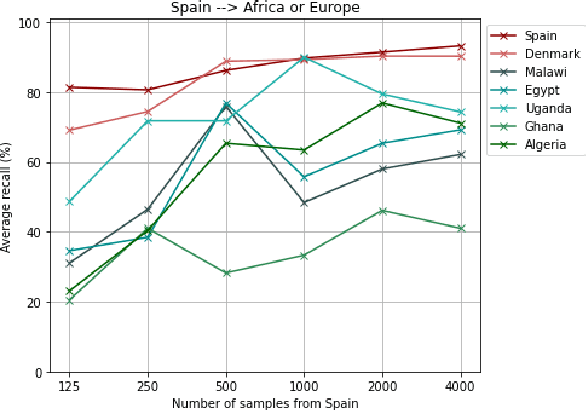
Abstract:Most artificial intelligence (AI) research have concentrated in high-income countries, where imaging data, IT infrastructures and clinical expertise are plentiful. However, slower progress has been made in limited-resource environments where medical imaging is needed. For example, in Sub-Saharan Africa the rate of perinatal mortality is very high due to limited access to antenatal screening. In these countries, AI models could be implemented to help clinicians acquire fetal ultrasound planes for diagnosis of fetal abnormalities. So far, deep learning models have been proposed to identify standard fetal planes, but there is no evidence of their ability to generalise in centres with limited access to high-end ultrasound equipment and data. This work investigates different strategies to reduce the domain-shift effect for a fetal plane classification model trained on a high-resource clinical centre and transferred to a new low-resource centre. To that end, a classifier trained with 1,792 patients from Spain is first evaluated on a new centre in Denmark in optimal conditions with 1,008 patients and is later optimised to reach the same performance in five African centres (Egypt, Algeria, Uganda, Ghana and Malawi) with 25 patients each. The results show that a transfer learning approach can be a solution to integrate small-size African samples with existing large-scale databases in developed countries. In particular, the model can be re-aligned and optimised to boost the performance on African populations by increasing the recall to $0.92 \pm 0.04$ and at the same time maintaining a high precision across centres. This framework shows promise for building new AI models generalisable across clinical centres with limited data acquired in challenging and heterogeneous conditions and calls for further research to develop new solutions for usability of AI in countries with less resources.
MixMOOD: A systematic approach to class distribution mismatch in semi-supervised learning using deep dataset dissimilarity measures
Jun 14, 2020



Abstract:In this work, we propose MixMOOD - a systematic approach to mitigate effect of class distribution mismatch in semi-supervised deep learning (SSDL) with MixMatch. This work is divided into two components: (i) an extensive out of distribution (OOD) ablation test bed for SSDL and (ii) a quantitative unlabelled dataset selection heuristic referred to as MixMOOD. In the first part, we analyze the sensitivity of MixMatch accuracy under 90 different distribution mismatch scenarios across three multi-class classification tasks. These are designed to systematically understand how OOD unlabelled data affects MixMatch performance. In the second part, we propose an efficient and effective method, called deep dataset dissimilarity measures (DeDiMs), to compare labelled and unlabelled datasets. The proposed DeDiMs are quick to evaluate and model agnostic. They use the feature space of a generic Wide-ResNet and can be applied prior to learning. Our test results reveal that supposed semantic similarity between labelled and unlabelled data is not a good heuristic for unlabelled data selection. In contrast, strong correlation between MixMatch accuracy and the proposed DeDiMs allow us to quantitatively rank different unlabelled datasets ante hoc according to expected MixMatch accuracy. This is what we call MixMOOD. Furthermore, we argue that the MixMOOD approach can aid to standardize the evaluation of different semi-supervised learning techniques under real world scenarios involving out of distribution data.
Breast Tumor Segmentation and Shape Classification in Mammograms using Generative Adversarial and Convolutional Neural Network
Oct 23, 2018



Abstract:Mammogram inspection in search of breast tumors is a tough assignment that radiologists must carry out frequently. Therefore, image analysis methods are needed for the detection and delineation of breast masses, which portray crucial morphological information that will support reliable diagnosis. In this paper, we proposed a conditional Generative Adversarial Network (cGAN) devised to segment a breast mass within a region of interest (ROI) in a mammogram. The generative network learns to recognize the breast mass area and to create the binary mask that outlines the breast mass. In turn, the adversarial network learns to distinguish between real (ground truth) and synthetic segmentations, thus enforcing the generative network to create binary masks as realistic as possible. The cGAN works well even when the number of training samples are limited. Therefore, the proposed method outperforms several state-of-the-art approaches. This hypothesis is corroborated by diverse experiments performed on two datasets, the public INbreast and a private in-house dataset. The proposed segmentation model provides a high Dice coefficient and Intersection over Union (IoU) of 94% and 87%, respectively. In addition, a shape descriptor based on a Convolutional Neural Network (CNN) is proposed to classify the generated masks into four mass shapes: irregular, lobular, oval and round. The proposed shape descriptor was trained on Digital Database for Screening Mammography (DDSM) yielding an overall accuracy of 80%, which outperforms the current state-of-the-art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge