Santiago Romani
An Efficient Solution for Breast Tumor Segmentation and Classification in Ultrasound Images Using Deep Adversarial Learning
Jul 01, 2019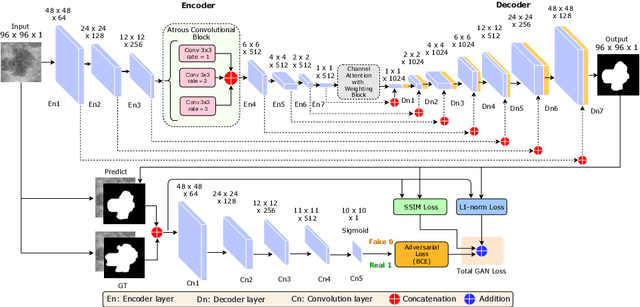
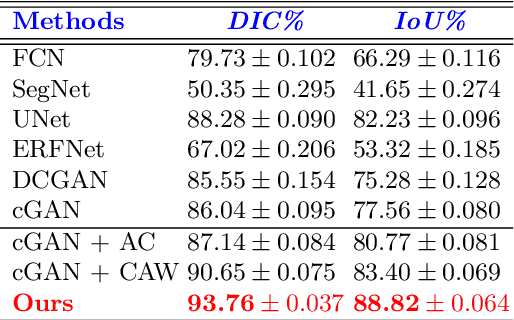
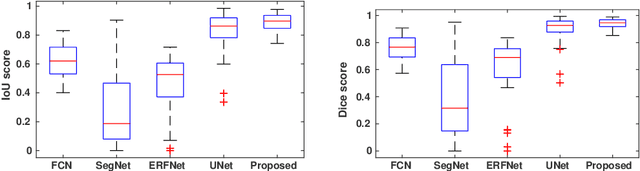
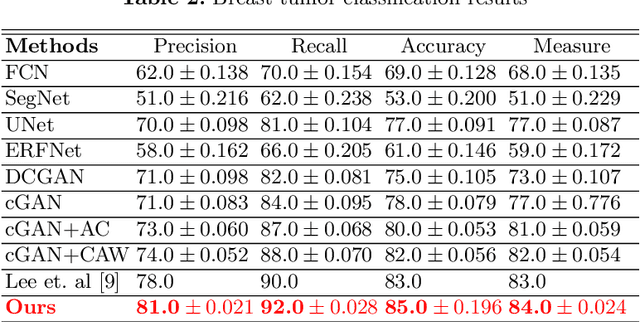
Abstract:This paper proposes an efficient solution for tumor segmentation and classification in breast ultrasound (BUS) images. We propose to add an atrous convolution layer to the conditional generative adversarial network (cGAN) segmentation model to learn tumor features at different resolutions of BUS images. To automatically re-balance the relative impact of each of the highest level encoded features, we also propose to add a channel-wise weighting block in the network. In addition, the SSIM and L1-norm loss with the typical adversarial loss are used as a loss function to train the model. Our model outperforms the state-of-the-art segmentation models in terms of the Dice and IoU metrics, achieving top scores of 93.76% and 88.82%, respectively. In the classification stage, we show that few statistics features extracted from the shape of the boundaries of the predicted masks can properly discriminate between benign and malignant tumors with an accuracy of 85%$
Breast Tumor Segmentation and Shape Classification in Mammograms using Generative Adversarial and Convolutional Neural Network
Oct 23, 2018



Abstract:Mammogram inspection in search of breast tumors is a tough assignment that radiologists must carry out frequently. Therefore, image analysis methods are needed for the detection and delineation of breast masses, which portray crucial morphological information that will support reliable diagnosis. In this paper, we proposed a conditional Generative Adversarial Network (cGAN) devised to segment a breast mass within a region of interest (ROI) in a mammogram. The generative network learns to recognize the breast mass area and to create the binary mask that outlines the breast mass. In turn, the adversarial network learns to distinguish between real (ground truth) and synthetic segmentations, thus enforcing the generative network to create binary masks as realistic as possible. The cGAN works well even when the number of training samples are limited. Therefore, the proposed method outperforms several state-of-the-art approaches. This hypothesis is corroborated by diverse experiments performed on two datasets, the public INbreast and a private in-house dataset. The proposed segmentation model provides a high Dice coefficient and Intersection over Union (IoU) of 94% and 87%, respectively. In addition, a shape descriptor based on a Convolutional Neural Network (CNN) is proposed to classify the generated masks into four mass shapes: irregular, lobular, oval and round. The proposed shape descriptor was trained on Digital Database for Screening Mammography (DDSM) yielding an overall accuracy of 80%, which outperforms the current state-of-the-art.
Retinal Optic Disc Segmentation using Conditional Generative Adversarial Network
Jun 11, 2018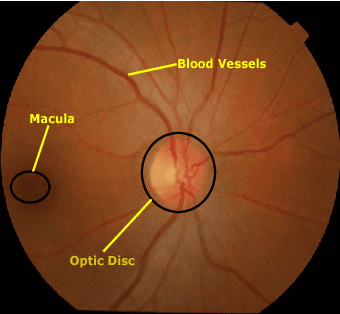
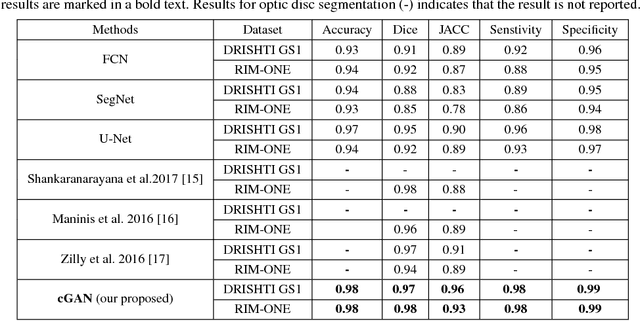
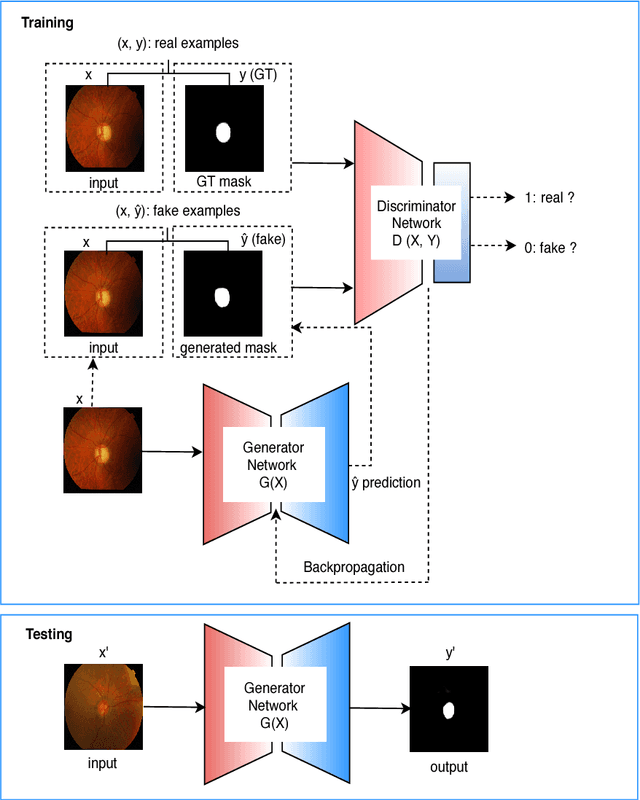

Abstract:This paper proposed a retinal image segmentation method based on conditional Generative Adversarial Network (cGAN) to segment optic disc. The proposed model consists of two successive networks: generator and discriminator. The generator learns to map information from the observing input (i.e., retinal fundus color image), to the output (i.e., binary mask). Then, the discriminator learns as a loss function to train this mapping by comparing the ground-truth and the predicted output with observing the input image as a condition.Experiments were performed on two publicly available dataset; DRISHTI GS1 and RIM-ONE. The proposed model outperformed state-of-the-art-methods by achieving around 0.96% and 0.98% of Jaccard and Dice coefficients, respectively. Moreover, an image segmentation is performed in less than a second on recent GPU.
Conditional Generative Adversarial and Convolutional Networks for X-ray Breast Mass Segmentation and Shape Classification
Jun 10, 2018


Abstract:This paper proposes a novel approach based on conditional Generative Adversarial Networks (cGAN) for breast mass segmentation in mammography. We hypothesized that the cGAN structure is well-suited to accurately outline the mass area, especially when the training data is limited. The generative network learns intrinsic features of tumors while the adversarial network enforces segmentations to be similar to the ground truth. Experiments performed on dozens of malignant tumors extracted from the public DDSM dataset and from our in-house private dataset confirm our hypothesis with very high Dice coefficient and Jaccard index (>94% and >89%, respectively) outperforming the scores obtained by other state-of-the-art approaches. Furthermore, in order to detect portray significant morphological features of the segmented tumor, a specific Convolutional Neural Network (CNN) have also been designed for classifying the segmented tumor areas into four types (irregular, lobular, oval and round), which provides an overall accuracy about 72% with the DDSM dataset.
SLSDeep: Skin Lesion Segmentation Based on Dilated Residual and Pyramid Pooling Networks
May 31, 2018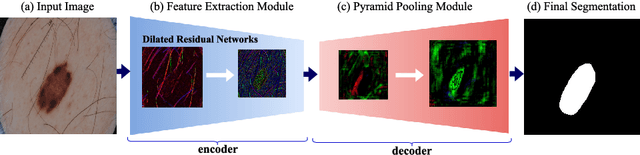
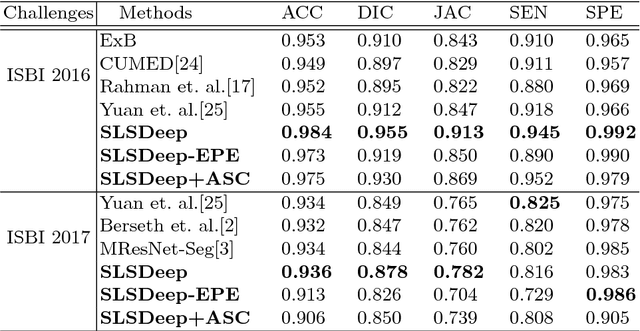
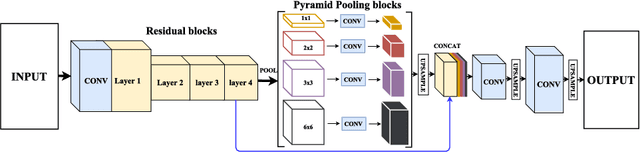
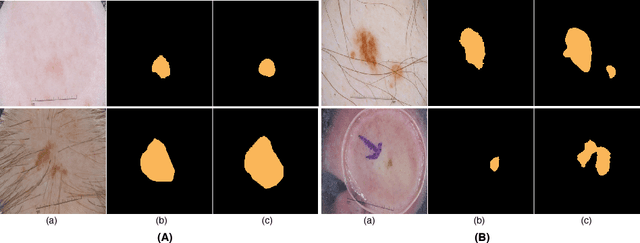
Abstract:Skin lesion segmentation (SLS) in dermoscopic images is a crucial task for automated diagnosis of melanoma. In this paper, we present a robust deep learning SLS model, so-called SLSDeep, which is represented as an encoder-decoder network. The encoder network is constructed by dilated residual layers, in turn, a pyramid pooling network followed by three convolution layers is used for the decoder. Unlike the traditional methods employing a cross-entropy loss, we investigated a loss function by combining both Negative Log Likelihood (NLL) and End Point Error (EPE) to accurately segment the melanoma regions with sharp boundaries. The robustness of the proposed model was evaluated on two public databases: ISBI 2016 and 2017 for skin lesion analysis towards melanoma detection challenge. The proposed model outperforms the state-of-the-art methods in terms of segmentation accuracy. Moreover, it is capable to segment more than $100$ images of size 384x384 per second on a recent GPU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge