Petia Radeva
BenchSeg: A Large-Scale Dataset and Benchmark for Multi-View Food Video Segmentation
Jan 12, 2026Abstract:Food image segmentation is a critical task for dietary analysis, enabling accurate estimation of food volume and nutrients. However, current methods suffer from limited multi-view data and poor generalization to new viewpoints. We introduce BenchSeg, a novel multi-view food video segmentation dataset and benchmark. BenchSeg aggregates 55 dish scenes (from Nutrition5k, Vegetables & Fruits, MetaFood3D, and FoodKit) with 25,284 meticulously annotated frames, capturing each dish under free 360° camera motion. We evaluate a diverse set of 20 state-of-the-art segmentation models (e.g., SAM-based, transformer, CNN, and large multimodal) on the existing FoodSeg103 dataset and evaluate them (alone and combined with video-memory modules) on BenchSeg. Quantitative and qualitative results demonstrate that while standard image segmenters degrade sharply under novel viewpoints, memory-augmented methods maintain temporal consistency across frames. Our best model based on a combination of SeTR-MLA+XMem2 outperforms prior work (e.g., improving over FoodMem by ~2.63% mAP), offering new insights into food segmentation and tracking for dietary analysis. We release BenchSeg to foster future research. The project page including the dataset annotations and the food segmentation models can be found at https://amughrabi.github.io/benchseg.
DADO: A Depth-Attention framework for Object Discovery
Oct 08, 2025Abstract:Unsupervised object discovery, the task of identifying and localizing objects in images without human-annotated labels, remains a significant challenge and a growing focus in computer vision. In this work, we introduce a novel model, DADO (Depth-Attention self-supervised technique for Discovering unseen Objects), which combines an attention mechanism and a depth model to identify potential objects in images. To address challenges such as noisy attention maps or complex scenes with varying depth planes, DADO employs dynamic weighting to adaptively emphasize attention or depth features based on the global characteristics of each image. We evaluated DADO on standard benchmarks, where it outperforms state-of-the-art methods in object discovery accuracy and robustness without the need for fine-tuning.
* 21st International Conference in Computer Analysis of Images and Patterns (CAIP 2025)
Advancing Image-Based Grapevine Variety Classification with a New Benchmark and Evaluation of Masked Autoencoders
Jun 16, 2025Abstract:Grapevine varieties are essential for the economies of many wine-producing countries, influencing the production of wine, juice, and the consumption of fruits and leaves. Traditional identification methods, such as ampelography and molecular analysis, have limitations: ampelography depends on expert knowledge and is inherently subjective, while molecular methods are costly and time-intensive. To address these limitations, recent studies have applied deep learning (DL) models to classify grapevine varieties using image data. However, due to the small dataset sizes, these methods often depend on transfer learning from datasets from other domains, e.g., ImageNet1K (IN1K), which can lead to performance degradation due to domain shift and supervision collapse. In this context, self-supervised learning (SSL) methods can be a good tool to avoid this performance degradation, since they can learn directly from data, without external labels. This study presents an evaluation of Masked Autoencoders (MAEs) for identifying grapevine varieties based on field-acquired images. The main contributions of this study include two benchmarks comprising 43 grapevine varieties collected across different seasons, an analysis of MAE's application in the agricultural context, and a performance comparison of trained models across seasons. Our results show that a ViT-B/16 model pre-trained with MAE and the unlabeled dataset achieved an F1 score of 0.7956, outperforming all other models. Additionally, we observed that pre-trained models benefit from long pre-training, perform well under low-data training regime, and that simple data augmentation methods are more effective than complex ones. The study also found that the mask ratio in MAE impacts performance only marginally.
Representation Discrepancy Bridging Method for Remote Sensing Image-Text Retrieval
May 22, 2025Abstract:Remote Sensing Image-Text Retrieval (RSITR) plays a critical role in geographic information interpretation, disaster monitoring, and urban planning by establishing semantic associations between image and textual descriptions. Existing Parameter-Efficient Fine-Tuning (PEFT) methods for Vision-and-Language Pre-training (VLP) models typically adopt symmetric adapter structures for exploring cross-modal correlations. However, the strong discriminative nature of text modality may dominate the optimization process and inhibits image representation learning. The nonnegligible imbalanced cross-modal optimization remains a bottleneck to enhancing the model performance. To address this issue, this study proposes a Representation Discrepancy Bridging (RDB) method for the RSITR task. On the one hand, a Cross-Modal Asymmetric Adapter (CMAA) is designed to enable modality-specific optimization and improve feature alignment. The CMAA comprises a Visual Enhancement Adapter (VEA) and a Text Semantic Adapter (TSA). VEA mines fine-grained image features by Differential Attention (DA) mechanism, while TSA identifies key textual semantics through Hierarchical Attention (HA) mechanism. On the other hand, this study extends the traditional single-task retrieval framework to a dual-task optimization framework and develops a Dual-Task Consistency Loss (DTCL). The DTCL improves cross-modal alignment robustness through an adaptive weighted combination of cross-modal, classification, and exponential moving average consistency constraints. Experiments on RSICD and RSITMD datasets show that the proposed RDB method achieves a 6%-11% improvement in mR metrics compared to state-of-the-art PEFT methods and a 1.15%-2% improvement over the full fine-tuned GeoRSCLIP model.
VolE: A Point-cloud Framework for Food 3D Reconstruction and Volume Estimation
May 15, 2025



Abstract:Accurate food volume estimation is crucial for medical nutrition management and health monitoring applications, but current food volume estimation methods are often limited by mononuclear data, leveraging single-purpose hardware such as 3D scanners, gathering sensor-oriented information such as depth information, or relying on camera calibration using a reference object. In this paper, we present VolE, a novel framework that leverages mobile device-driven 3D reconstruction to estimate food volume. VolE captures images and camera locations in free motion to generate precise 3D models, thanks to AR-capable mobile devices. To achieve real-world measurement, VolE is a reference- and depth-free framework that leverages food video segmentation for food mask generation. We also introduce a new food dataset encompassing the challenging scenarios absent in the previous benchmarks. Our experiments demonstrate that VolE outperforms the existing volume estimation techniques across multiple datasets by achieving 2.22 % MAPE, highlighting its superior performance in food volume estimation.
Calibration and Uncertainty for multiRater Volume Assessment in multiorgan Segmentation (CURVAS) challenge results
May 13, 2025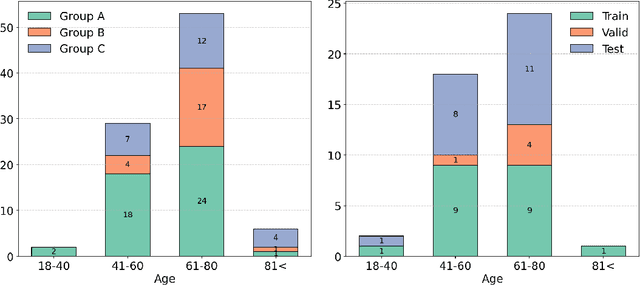
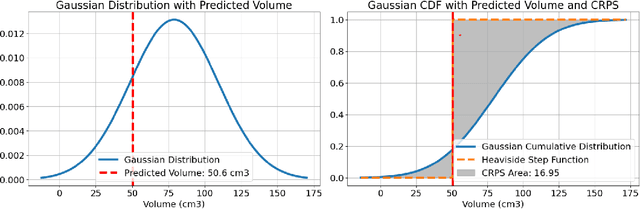

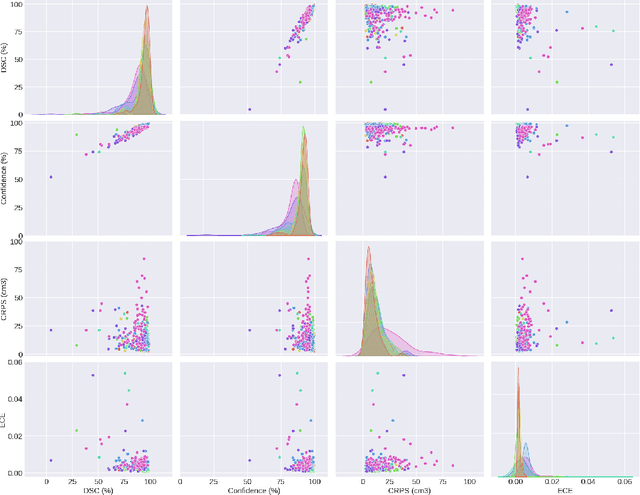
Abstract:Deep learning (DL) has become the dominant approach for medical image segmentation, yet ensuring the reliability and clinical applicability of these models requires addressing key challenges such as annotation variability, calibration, and uncertainty estimation. This is why we created the Calibration and Uncertainty for multiRater Volume Assessment in multiorgan Segmentation (CURVAS), which highlights the critical role of multiple annotators in establishing a more comprehensive ground truth, emphasizing that segmentation is inherently subjective and that leveraging inter-annotator variability is essential for robust model evaluation. Seven teams participated in the challenge, submitting a variety of DL models evaluated using metrics such as Dice Similarity Coefficient (DSC), Expected Calibration Error (ECE), and Continuous Ranked Probability Score (CRPS). By incorporating consensus and dissensus ground truth, we assess how DL models handle uncertainty and whether their confidence estimates align with true segmentation performance. Our findings reinforce the importance of well-calibrated models, as better calibration is strongly correlated with the quality of the results. Furthermore, we demonstrate that segmentation models trained on diverse datasets and enriched with pre-trained knowledge exhibit greater robustness, particularly in cases deviating from standard anatomical structures. Notably, the best-performing models achieved high DSC and well-calibrated uncertainty estimates. This work underscores the need for multi-annotator ground truth, thorough calibration assessments, and uncertainty-aware evaluations to develop trustworthy and clinically reliable DL-based medical image segmentation models.
Conjuring Positive Pairs for Efficient Unification of Representation Learning and Image Synthesis
Mar 20, 2025Abstract:While representation learning and generative modeling seek to understand visual data, unifying both domains remains unexplored. Recent Unified Self-Supervised Learning (SSL) methods have started to bridge the gap between both paradigms. However, they rely solely on semantic token reconstruction, which requires an external tokenizer during training -- introducing a significant overhead. In this work, we introduce Sorcen, a novel unified SSL framework, incorporating a synergic Contrastive-Reconstruction objective. Our Contrastive objective, "Echo Contrast", leverages the generative capabilities of Sorcen, eliminating the need for additional image crops or augmentations during training. Sorcen "generates" an echo sample in the semantic token space, forming the contrastive positive pair. Sorcen operates exclusively on precomputed tokens, eliminating the need for an online token transformation during training, thereby significantly reducing computational overhead. Extensive experiments on ImageNet-1k demonstrate that Sorcen outperforms the previous Unified SSL SoTA by 0.4%, 1.48 FID, 1.76%, and 1.53% on linear probing, unconditional image generation, few-shot learning, and transfer learning, respectively, while being 60.8% more efficient. Additionally, Sorcen surpasses previous single-crop MIM SoTA in linear probing and achieves SoTA performance in unconditional image generation, highlighting significant improvements and breakthroughs in Unified SSL models.
Multi-label out-of-distribution detection via evidential learning
Feb 25, 2025



Abstract:A crucial requirement for machine learning algorithms is not only to perform well, but also to show robustness and adaptability when encountering novel scenarios. One way to achieve these characteristics is to endow the deep learning models with the ability to detect out-of-distribution (OOD) data, i.e. data that belong to distributions different from the one used during their training. It is even a more complicated situation, when these data usually are multi-label. In this paper, we propose an approach based on evidential deep learning in order to meet these challenges applied to visual recognition problems. More concretely, we designed a CNN architecture that uses a Beta Evidential Neural Network to compute both the likelihood and the predictive uncertainty of the samples. Based on these results, we propose afterwards two new uncertainty-based scores for OOD data detection: (i) OOD - score Max, based on the maximum evidence; and (ii) OOD score - Sum, which considers the evidence from all outputs. Extensive experiments have been carried out to validate the proposed approach using three widely-used datasets: PASCAL-VOC, MS-COCO and NUS-WIDE, demonstrating its outperformance over several State-of-the-Art methods.
Explainable AI model reveals disease-related mechanisms in single-cell RNA-seq data
Jan 07, 2025Abstract:Neurodegenerative diseases (NDDs) are complex and lack effective treatment due to their poorly understood mechanism. The increasingly used data analysis from Single nucleus RNA Sequencing (snRNA-seq) allows to explore transcriptomic events at a single cell level, yet face challenges in interpreting the mechanisms underlying a disease. On the other hand, Neural Network (NN) models can handle complex data to offer insights but can be seen as black boxes with poor interpretability. In this context, explainable AI (XAI) emerges as a solution that could help to understand disease-associated mechanisms when combined with efficient NN models. However, limited research explores XAI in single-cell data. In this work, we implement a method for identifying disease-related genes and the mechanistic explanation of disease progression based on NN model combined with SHAP. We analyze available Huntington's disease (HD) data to identify both HD-altered genes and mechanisms by adding Gene Set Enrichment Analysis (GSEA) comparing two methods, differential gene expression analysis (DGE) and NN combined with SHAP approach. Our results show that DGE and SHAP approaches offer both common and differential sets of altered genes and pathways, reinforcing the usefulness of XAI methods for a broader perspective of disease.
Hybrid deep learning-based strategy for the hepatocellular carcinoma cancer grade classification of H&E stained liver histopathology images
Dec 04, 2024



Abstract:Hepatocellular carcinoma (HCC) is a common type of liver cancer whose early-stage diagnosis is a common challenge, mainly due to the manual assessment of hematoxylin and eosin-stained whole slide images, which is a time-consuming process and may lead to variability in decision-making. For accurate detection of HCC, we propose a hybrid deep learning-based architecture that uses transfer learning to extract the features from pre-trained convolutional neural network (CNN) models and a classifier made up of a sequence of fully connected layers. This study uses a publicly available The Cancer Genome Atlas Hepatocellular Carcinoma (TCGA-LIHC)database (n=491) for model development and database of Kasturba Gandhi Medical College (KMC), India for validation. The pre-processing step involves patch extraction, colour normalization, and augmentation that results in 3920 patches for the TCGA dataset. The developed hybrid deep neural network consisting of a CNN-based pre-trained feature extractor and a customized artificial neural network-based classifier is trained using five-fold cross-validation. For this study, eight different state-of-the-art models are trained and tested as feature extractors for the proposed hybrid model. The proposed hybrid model with ResNet50-based feature extractor provided the sensitivity, specificity, F1-score, accuracy, and AUC of 100.00%, 100.00%, 100.00%, 100.00%, and 1.00, respectively on the TCGA database. On the KMC database, EfficientNetb3 resulted in the optimal choice of the feature extractor giving sensitivity, specificity, F1-score, accuracy, and AUC of 96.97, 98.85, 96.71, 96.71, and 0.99, respectively. The proposed hybrid models showed improvement in accuracy of 2% and 4% over the pre-trained models in TCGA-LIHC and KMC databases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge