Alexander Wong
DarwinAI, University of Waterloo
Understanding vision transformer robustness through the lens of out-of-distribution detection
Feb 01, 2026Abstract:Vision transformers have shown remarkable performance in vision tasks, but enabling them for accessible and real-time use is still challenging. Quantization reduces memory and inference costs at the risk of performance loss. Strides have been made to mitigate low precision issues mainly by understanding in-distribution (ID) task behaviour, but the attention mechanism may provide insight on quantization attributes by exploring out-of-distribution (OOD) situations. We investigate the behaviour of quantized small-variant popular vision transformers (DeiT, DeiT3, and ViT) on common OOD datasets. ID analyses show the initial instabilities of 4-bit models, particularly of those trained on the larger ImageNet-22k, as the strongest FP32 model, DeiT3, sharply drop 17% from quantization error to be one of the weakest 4-bit models. While ViT shows reasonable quantization robustness for ID calibration, OOD detection reveals more: ViT and DeiT3 pretrained on ImageNet-22k respectively experienced a 15.0% and 19.2% average quantization delta in AUPR-out between full precision to 4-bit while their ImageNet-1k-only counterparts experienced a 9.5% and 12.0% delta. Overall, our results suggest pretraining on large scale datasets may hinder low-bit quantization robustness in OOD detection and that data augmentation may be a more beneficial option.
Composite Classifier-Free Guidance for Multi-Modal Conditioning in Wind Dynamics Super-Resolution
Dec 13, 2025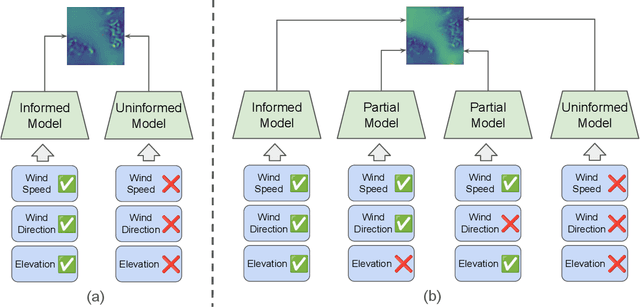
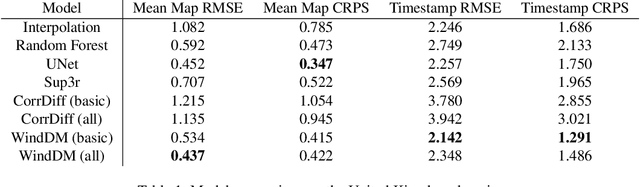
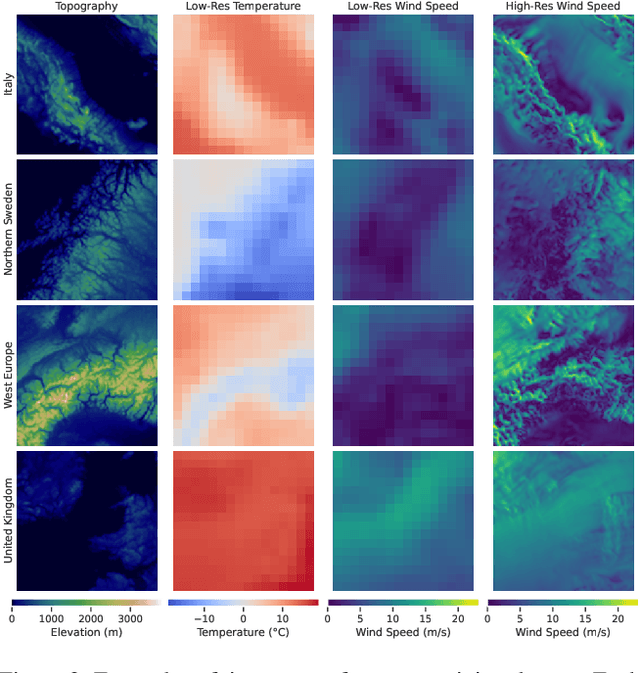
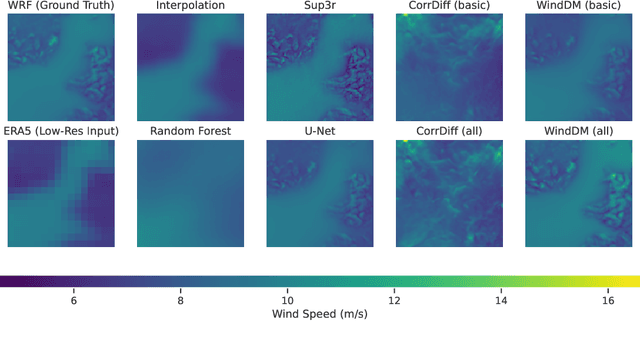
Abstract:Various weather modelling problems (e.g., weather forecasting, optimizing turbine placements, etc.) require ample access to high-resolution, highly accurate wind data. Acquiring such high-resolution wind data, however, remains a challenging and expensive endeavour. Traditional reconstruction approaches are typically either cost-effective or accurate, but not both. Deep learning methods, including diffusion models, have been proposed to resolve this trade-off by leveraging advances in natural image super-resolution. Wind data, however, is distinct from natural images, and wind super-resolvers often use upwards of 10 input channels, significantly more than the usual 3-channel RGB inputs in natural images. To better leverage a large number of conditioning variables in diffusion models, we present a generalization of classifier-free guidance (CFG) to multiple conditioning inputs. Our novel composite classifier-free guidance (CCFG) can be dropped into any pre-trained diffusion model trained with standard CFG dropout. We demonstrate that CCFG outputs are higher-fidelity than those from CFG on wind super-resolution tasks. We present WindDM, a diffusion model trained for industrial-scale wind dynamics reconstruction and leveraging CCFG. WindDM achieves state-of-the-art reconstruction quality among deep learning models and costs up to $1000\times$ less than classical methods.
SCALEX: Scalable Concept and Latent Exploration for Diffusion Models
Nov 13, 2025Abstract:Image generation models frequently encode social biases, including stereotypes tied to gender, race, and profession. Existing methods for analyzing these biases in diffusion models either focus narrowly on predefined categories or depend on manual interpretation of latent directions. These constraints limit scalability and hinder the discovery of subtle or unanticipated patterns. We introduce SCALEX, a framework for scalable and automated exploration of diffusion model latent spaces. SCALEX extracts semantically meaningful directions from H-space using only natural language prompts, enabling zero-shot interpretation without retraining or labelling. This allows systematic comparison across arbitrary concepts and large-scale discovery of internal model associations. We show that SCALEX detects gender bias in profession prompts, ranks semantic alignment across identity descriptors, and reveals clustered conceptual structure without supervision. By linking prompts to latent directions directly, SCALEX makes bias analysis in diffusion models more scalable, interpretable, and extensible than prior approaches.
Cancer-Net PCa-MultiSeg: Multimodal Enhancement of Prostate Cancer Lesion Segmentation Using Synthetic Correlated Diffusion Imaging
Nov 11, 2025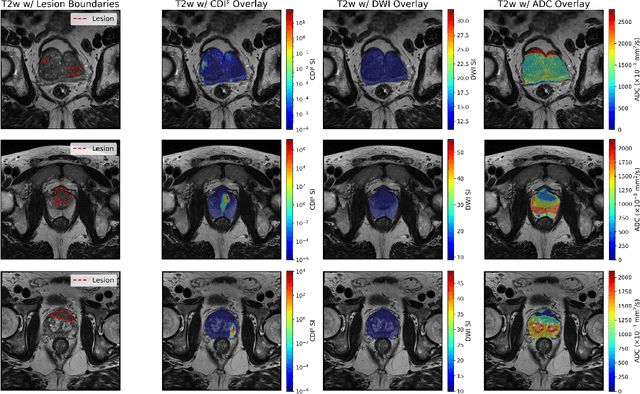
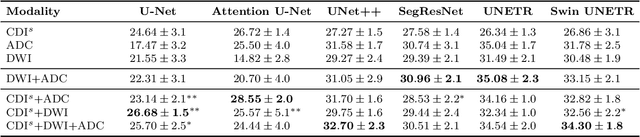
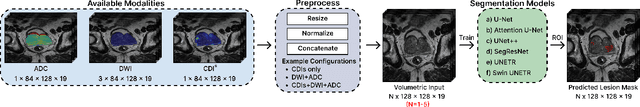
Abstract:Current deep learning approaches for prostate cancer lesion segmentation achieve limited performance, with Dice scores of 0.32 or lower in large patient cohorts. To address this limitation, we investigate synthetic correlated diffusion imaging (CDI$^s$) as an enhancement to standard diffusion-based protocols. We conduct a comprehensive evaluation across six state-of-the-art segmentation architectures using 200 patients with co-registered CDI$^s$, diffusion-weighted imaging (DWI) and apparent diffusion coefficient (ADC) sequences. We demonstrate that CDI$^s$ integration reliably enhances or preserves segmentation performance in 94% of evaluated configurations, with individual architectures achieving up to 72.5% statistically significant relative improvement over baseline modalities. CDI$^s$ + DWI emerges as the safest enhancement pathway, achieving significant improvements in half of evaluated architectures with zero instances of degradation. Since CDI$^s$ derives from existing DWI acquisitions without requiring additional scan time or architectural modifications, it enables immediate deployment in clinical workflows. Our results establish validated integration pathways for CDI$^s$ as a practical drop-in enhancement for PCa lesion segmentation tasks across diverse deep learning architectures.
An Explainable Hybrid AI Framework for Enhanced Tuberculosis and Symptom Detection
Oct 21, 2025



Abstract:Tuberculosis remains a critical global health issue, particularly in resource-limited and remote areas. Early detection is vital for treatment, yet the lack of skilled radiologists underscores the need for artificial intelligence (AI)-driven screening tools. Developing reliable AI models is challenging due to the necessity for large, high-quality datasets, which are costly to obtain. To tackle this, we propose a teacher--student framework which enhances both disease and symptom detection on chest X-rays by integrating two supervised heads and a self-supervised head. Our model achieves an accuracy of 98.85% for distinguishing between COVID-19, tuberculosis, and normal cases, and a macro-F1 score of 90.09% for multilabel symptom detection, significantly outperforming baselines. The explainability assessments also show the model bases its predictions on relevant anatomical features, demonstrating promise for deployment in clinical screening and triage settings.
Starting Positions Matter: A Study on Better Weight Initialization for Neural Network Quantization
Jun 12, 2025



Abstract:Deep neural network (DNN) quantization for fast, efficient inference has been an important tool in limiting the cost of machine learning (ML) model inference. Quantization-specific model development techniques such as regularization, quantization-aware training, and quantization-robustness penalties have served to greatly boost the accuracy and robustness of modern DNNs. However, very little exploration has been done on improving the initial conditions of DNN training for quantization. Just as random weight initialization has been shown to significantly impact test accuracy of floating point models, it would make sense that different weight initialization methods impact quantization robustness of trained models. We present an extensive study examining the effects of different weight initializations on a variety of CNN building blocks commonly used in efficient CNNs. This analysis reveals that even with varying CNN architectures, the choice of random weight initializer can significantly affect final quantization robustness. Next, we explore a new method for quantization-robust CNN initialization -- using Graph Hypernetworks (GHN) to predict parameters of quantized DNNs. Besides showing that GHN-predicted parameters are quantization-robust after regular float32 pretraining (of the GHN), we find that finetuning GHNs to predict parameters for quantized graphs (which we call GHN-QAT) can further improve quantized accuracy of CNNs. Notably, GHN-QAT shows significant accuracy improvements for even 4-bit quantization and better-than-random accuracy for 2-bits. To the best of our knowledge, this is the first in-depth study on quantization-aware DNN weight initialization. GHN-QAT offers a novel approach to quantized DNN model design. Future investigations, such as using GHN-QAT-initialized parameters for quantization-aware training, can further streamline the DNN quantization process.
SAMJAM: Zero-Shot Video Scene Graph Generation for Egocentric Kitchen Videos
Apr 10, 2025Abstract:Video Scene Graph Generation (VidSGG) is an important topic in understanding dynamic kitchen environments. Current models for VidSGG require extensive training to produce scene graphs. Recently, Vision Language Models (VLM) and Vision Foundation Models (VFM) have demonstrated impressive zero-shot capabilities in a variety of tasks. However, VLMs like Gemini struggle with the dynamics for VidSGG, failing to maintain stable object identities across frames. To overcome this limitation, we propose SAMJAM, a zero-shot pipeline that combines SAM2's temporal tracking with Gemini's semantic understanding. SAM2 also improves upon Gemini's object grounding by producing more accurate bounding boxes. In our method, we first prompt Gemini to generate a frame-level scene graph. Then, we employ a matching algorithm to map each object in the scene graph with a SAM2-generated or SAM2-propagated mask, producing a temporally-consistent scene graph in dynamic environments. Finally, we repeat this process again in each of the following frames. We empirically demonstrate that SAMJAM outperforms Gemini by 8.33% in mean recall on the EPIC-KITCHENS and EPIC-KITCHENS-100 datasets.
The Efficacy of Semantics-Preserving Transformations in Self-Supervised Learning for Medical Ultrasound
Apr 10, 2025Abstract:Data augmentation is a central component of joint embedding self-supervised learning (SSL). Approaches that work for natural images may not always be effective in medical imaging tasks. This study systematically investigated the impact of data augmentation and preprocessing strategies in SSL for lung ultrasound. Three data augmentation pipelines were assessed: (1) a baseline pipeline commonly used across imaging domains, (2) a novel semantic-preserving pipeline designed for ultrasound, and (3) a distilled set of the most effective transformations from both pipelines. Pretrained models were evaluated on multiple classification tasks: B-line detection, pleural effusion detection, and COVID-19 classification. Experiments revealed that semantics-preserving data augmentation resulted in the greatest performance for COVID-19 classification - a diagnostic task requiring global image context. Cropping-based methods yielded the greatest performance on the B-line and pleural effusion object classification tasks, which require strong local pattern recognition. Lastly, semantics-preserving ultrasound image preprocessing resulted in increased downstream performance for multiple tasks. Guidance regarding data augmentation and preprocessing strategies was synthesized for practitioners working with SSL in ultrasound.
LangDA: Building Context-Awareness via Language for Domain Adaptive Semantic Segmentation
Mar 17, 2025Abstract:Unsupervised domain adaptation for semantic segmentation (DASS) aims to transfer knowledge from a label-rich source domain to a target domain with no labels. Two key approaches in DASS are (1) vision-only approaches using masking or multi-resolution crops, and (2) language-based approaches that use generic class-wise prompts informed by target domain (e.g. "a {snowy} photo of a {class}"). However, the former is susceptible to noisy pseudo-labels that are biased to the source domain. The latter does not fully capture the intricate spatial relationships of objects -- key for dense prediction tasks. To this end, we propose LangDA. LangDA addresses these challenges by, first, learning contextual relationships between objects via VLM-generated scene descriptions (e.g. "a pedestrian is on the sidewalk, and the street is lined with buildings."). Second, LangDA aligns the entire image features with text representation of this context-aware scene caption and learns generalized representations via text. With this, LangDA sets the new state-of-the-art across three DASS benchmarks, outperforming existing methods by 2.6%, 1.4% and 3.9%.
Cancer-Net PCa-Seg: Benchmarking Deep Learning Models for Prostate Cancer Segmentation Using Synthetic Correlated Diffusion Imaging
Jan 15, 2025


Abstract:Prostate cancer (PCa) is the most prevalent cancer among men in the United States, accounting for nearly 300,000 cases, 29% of all diagnoses and 35,000 total deaths in 2024. Traditional screening methods such as prostate-specific antigen (PSA) testing and magnetic resonance imaging (MRI) have been pivotal in diagnosis, but have faced limitations in specificity and generalizability. In this paper, we explore the potential of enhancing PCa lesion segmentation using a novel MRI modality called synthetic correlated diffusion imaging (CDI$^s$). We employ several state-of-the-art deep learning models, including U-Net, SegResNet, Swin UNETR, Attention U-Net, and LightM-UNet, to segment PCa lesions from a 200 CDI$^s$ patient cohort. We find that SegResNet achieved superior segmentation performance with a Dice-Sorensen coefficient (DSC) of $76.68 \pm 0.8$. Notably, the Attention U-Net, while slightly less accurate (DSC $74.82 \pm 2.0$), offered a favorable balance between accuracy and computational efficiency. Our findings demonstrate the potential of deep learning models in improving PCa lesion segmentation using CDI$^s$ to enhance PCa management and clinical support.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge